Command Palette
Search for a command to run...
Based on Billions of Genes From One Million Species, NVIDIA and Others Have Built the EDEN Series of Models, Achieving state-of-the-art (SOTA) Genome and Protein Prediction capabilities.

The fundamental goal of programmable biology is to achieve rational design and precise control of living systems, thereby bringing revolutionary therapies to complex diseases. However,This process has long been constrained by the inherent complexity of biological systems.Cross-scale regulatory networks, hidden long-sequence dependencies, and the diverse adaptability of organisms to environmental changes have all led to a dilemma of customization, low throughput, and high cost in traditional "trial and error" research and development.
Ultimately, the training data upon which current computational models rely, in terms of both scale and diversity, falls far short of encompassing the vast design space that life has developed over billions of years of evolution. These models therefore struggle to capture universal design principles.When faced with innovative therapy designs that are multimodal and cross-scale, the ability to generalize is severely lacking.
To overcome this fundamental limitation,Basecamp Research, NVIDIA, and several leading academic institutions jointly developed the EDEN series of metagenomic basic models.By learning from massive amounts of natural evolutionary data that cross species and relate to environmental information, the deep "grammar" and universal principles of biological design have been systematically extracted for the first time. The model has 28 billion parameters and has achieved state-of-the-art results in multiple benchmark tests. Its core breakthrough lies in its outstanding ability to understand and generate cross-species sequences, thus advancing bioengineering from "screening" to a new stage of "predictable programming".
To validate EDEN's capabilities as a unified biodesign engine, the research team conducted systematic tests across multiple treatment modalities. In gene therapy, EDEN, using only a 30-base hint at a target site, can de novo design of active recombinases capable of precisely integrating large fragments into the human genome. Regarding antimicrobial peptide design…The peptide library generated by the same model showed up to 97% activity against multidrug-resistant pathogens.It also possesses micromolar-level potency. At the ecosystem level, EDEN has successfully constructed a synthetic microbiome containing tens of thousands of artificial genomes, accurate metabolic pathways, and reasonable species relationships.
The related research findings, titled "Designing AI-programmable therapeutics with the EDEN family of foundation models," have been published as a preprint on bioRxiv.
Research highlights:
* It pioneered a new paradigm for learning universal design principles directly from evolutionary history, and through training with BaseData, a metagenomic database covering global biodiversity, it achieved outstanding cross-species sequence understanding and generation capabilities.
* This study validates the powerful versatility of a single fundamental model in driving multi-scale, multimodal therapy design, demonstrating that a single model can uniformly address complex design challenges ranging from molecules to ecosystems.
* EDEN, using only DNA clues, can design functional recombinases for multiple disease-related sites, achieving a functional hit rate of 63.21 TP3T at untrained targets.

Paper address:
https://doi.org/10.64898/2026.01.12.699009
Follow our official WeChat account and reply "EDEN" in the background to get the full PDF.
More AI frontier papers:
https://hyper.ai/papers
BaseData dataset: Reshaping biological AI data benchmarks with high-quality long sequences.
The BaseData dataset used in this study fundamentally breaks through the limitations of traditional biological databases. Traditional databases typically rely on limited reference genomes and fragmented short sequences, while BaseData aims to systematically capture complete evolutionary signals, constructing an evolutionary genomic data supply chain covering global biodiversity.
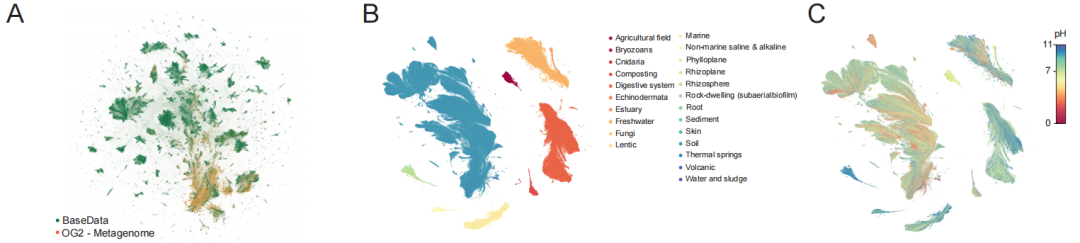
BaseData's core value is primarily reflected in its scale and strategic composition. As shown in the diagram below,It contains 9.7 trillion nucleotide markers for training, covering more than 1 million new species and 100 billion new genes.More importantly, the data is not randomly collected, but deliberately enriched with high-information-density sequences such as environmental metagenomics, bacteriophages, and mobile genetic elements. These data naturally record key evolutionary dynamics such as bacteriophage-host interactions and horizontal gene transfer, providing core material for models to learn universal functional rules across species.
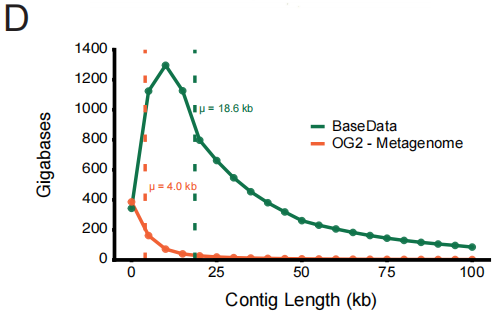
In terms of data quality, BaseData achieves a qualitative improvement, primarily in the completeness of the sequence context. Compared to the widely used OpenGenome-2 (OG2), its median length of contiguous sequence fragments (overlaps) reaches 18.6 kbp (OG2 is 4.0 kbp), and each assembly contains a significantly larger number of genes. This longer contiguous background is crucial for the model to understand intergenetic regulation and metabolic pathways.
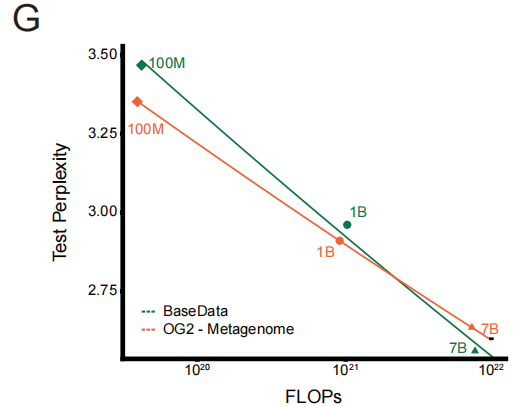
To quantify this quality advantage, the research team conducted a controlled experiment: training a series of models on equal-sized datasets from BaseData and OG2. The results clearly validated the "quality-aware scaling law." Under the same computational overhead, the model trained on BaseData experienced a faster decrease in test perplexity. A key finding is that large models (e.g., 7 billion parameters) can fully utilize the long-sequence information of BaseData, ultimately outperforming similar models trained on OG2.This directly demonstrates the decisive impact of long-range context on model performance.
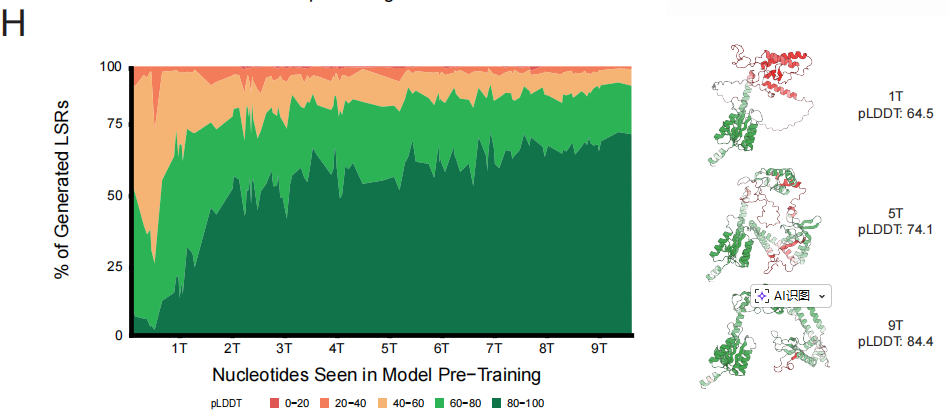
Based on this patternThe research team trained the EDEN-28B model with 28 billion parameters using the complete BaseData.This model not only achieved the lowest test perplexity, but its performance improvement trajectory also perfectly matches the scaling predictions derived from small-scale models. In downstream task monitoring, the structural confidence index of proteins generated by the model during pre-training showed a continuous monotonic increase with the training process, demonstrating that high-quality data directly and steadily improves the ability to generate data for practical treatment.
Furthermore, all data was obtained through standardized legal agreements covering 28 countries and 208 licenses, establishing a traceability and benefit-sharing framework from source to use, and setting necessary ethical and governance standards for large-scale biological AI research.
EDEN, a general-purpose biological design engine
The EDEN model family is designed with "scalability, universality and scalability" as its core design principles, with model parameters ranging from 100 million to 28 billion.Among them, EDEN-28B, as the core working model, has an architecture and training strategy that are deeply adapted to the unique properties of metagenomic data.
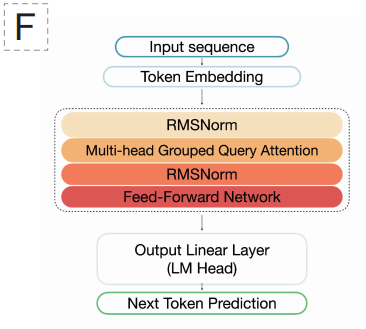
In terms of model architecture,EDEN adopts a decoder-only Transformer architecture validated by large-scale language models, specifically based on the design style of Llama 3.1.This choice is made possible by the Transformer's superior ability to model long-range dependencies. EDEN-28B comprises a 48-layer network with 6,144 hidden layers, 48 attention heads, and uses the SwiGLU activation function with RoPE positional encoding. The model employs a single-nucleotide resolution tokenization method with a vocabulary size of 512, enabling it to understand and generate DNA sequences at the most basic "letter" level.
A key technological highlight lies in its ability to generate long sequences. Although the model's context window is set to 8,192 labels, in practical applications,It can stably generate and accurately assemble coherent genome sequences of more than 13,000 base pairs while maintaining the correct gene order, reading frames, and regulatory element structures.This indicates that the model learns far more than local pattern matching; it can infer and apply a deeper "grammar" of genomic organization that goes beyond the physical window length. The entire training was completed on 1,008 H100 GPUs, achieving efficient learning from massive amounts of evolutionary data through large-scale distributed computing.
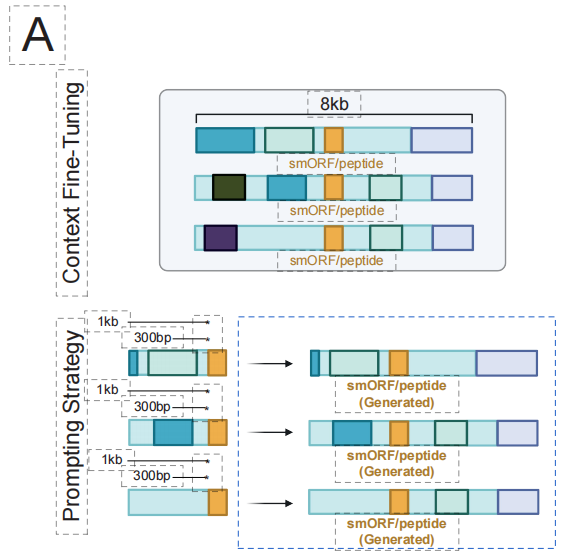
EDEN's core design philosophy follows a "pre-training-fine-tuning" paradigm. In the first stage, the model is pre-trained on a large scale on BaseData that covers evolutionary history across species, thereby internalizing general principles of biological design such as protein folding and metabolic pathway assembly.
Building on this solid foundation, we can then target specific therapeutic design tasks—such as designing recombinases that target specific DNA sites or generating novel antimicrobial peptides.With only a small amount of high-quality task-paired data for lightweight fine-tuning, the model can quickly master the "dialect" of the task.This design enables a single EDEN model to serve as a universal "biological sequence engine," flexibly adapting to and driving diverse therapeutic modalities ranging from gene insertion and peptide design to microbiome engineering, truly realizing the programmable biology vision of "one model, multiple capabilities."
Driving therapeutic innovation from the molecular, cellular to ecosystem levels
To systematically verify the universality and effectiveness of the EDEN model in actual treatment design, the research team selected four key directions that are very different in scale, pattern and biological complexity for experimental verification.
In the field of AI programmable gene insertion (aiPGI), the team has focused on overcoming the long-standing bottleneck of "precise integration of large DNA fragments".Traditional CRISPR technology relies on creating double-strand breaks, and natural large serine recombinases cannot recognize human genome sequences. As shown in the figure below, EDEN's solution is to construct the EDEN-LSR model, which can understand the mapping relationship of "target DNA sequence → corresponding recombinase," by fine-tuning the millions of LSR-attachment site pairings contained in the model.
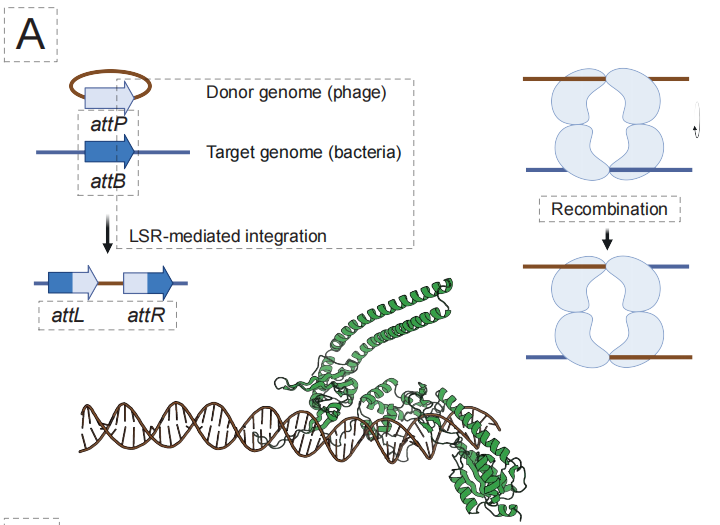
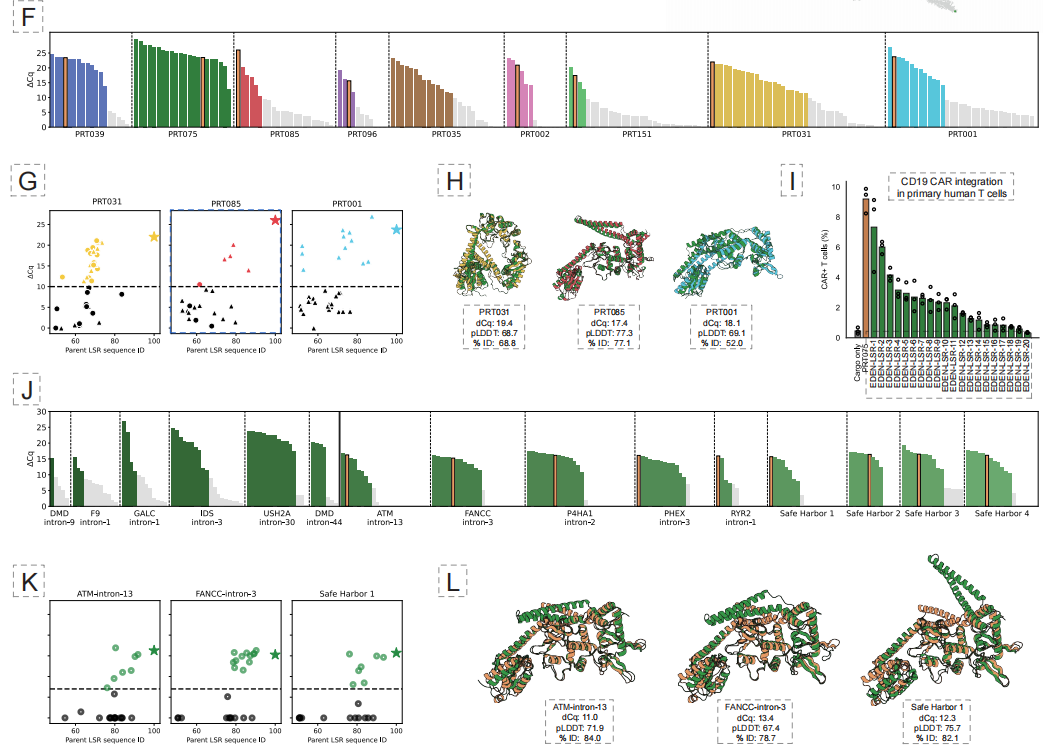
Experimental results showed that this approach successfully generated active LSRs for 10 different disease-related gene loci and 4 potential "safe harbor" loci, with an overall functional hit rate of 53.61 TP3T. More importantly,The 50% design enzyme can achieve treatment-related CAR gene insertion in primary human T cells, and some variants have achieved integration efficiencies of up to 40% in cell lines.This demonstrates its potential for clinical application.
In the field of novel bridging recombinases (BRs),The capabilities of the EDEN model have been further extended to a more programmable gene editing system—bridging recombinase.As shown in the figure below, in order to optimize the design, the team built the EDEN-BR-specific model by fine-tuning the model on millions of BR-containing genomic regions.
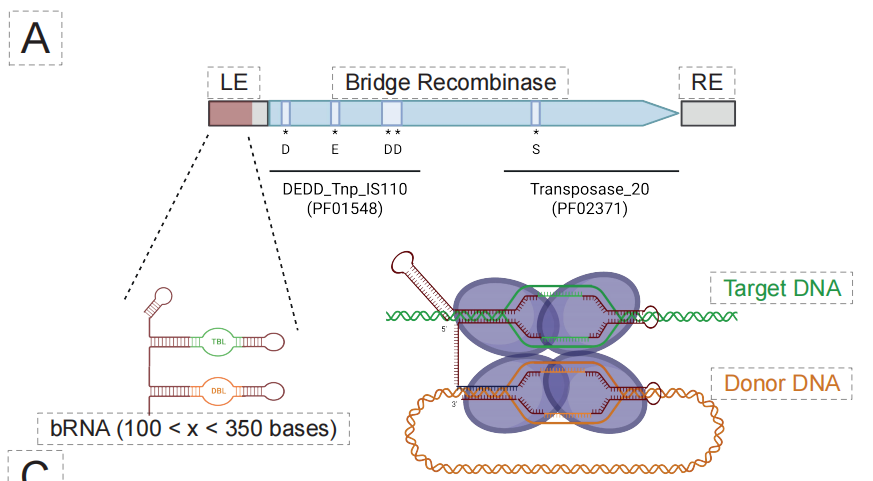
Key biochemical experiments validated the feasibility of this design process. As shown in the figure below, in preliminary cell-free testing, two of the 49 candidate sequences generated by EDEN-BR were confirmed to have definite recombinase activity. These two artificially designed proteins, named DF3843 and DF3881, have a maximum similarity of only 851 TP3T and 65.81 TP3T, respectively, to any known natural BR sequence. Their sequence similarity to a well-studied reference protein, ISCro4, is even lower than 351 TP3T, but they are highly similar in three-dimensional structure.This proves that EDEN does not simply mimic sequences, but rather has mastered the core structural logic that determines protein function and folding.
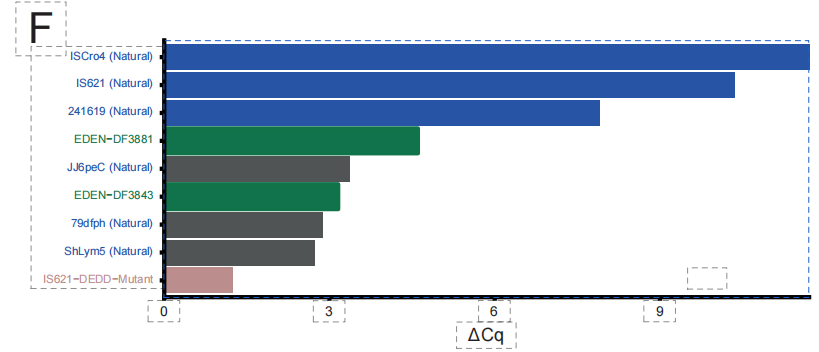
In the field of novel antimicrobial peptides (AMPs), the research team validated EDEN's ability to design novel antimicrobial peptides. As shown in the figure below,By employing a fine-tuning strategy that incorporates genomic contextual information, the model is able to generate novel antimicrobial peptide sequences.
The experimental verification has yielded groundbreaking results. As shown in the figure below,In an AMP library consisting of 33 generating peptides, up to 971 TP3T sequences exhibited antibacterial activity.Among them, for multidrug-resistant Gram-negative bacteria (such as Acinetobacter baumannii), the top-designed candidate achieved antibacterial concentrations at the micromolar level, demonstrating a strong ability to penetrate the outer membrane. These generated sequences generally showed low similarity to known databases, confirming that the model can overcome traditional homology limitations and achieve true "de novo design".
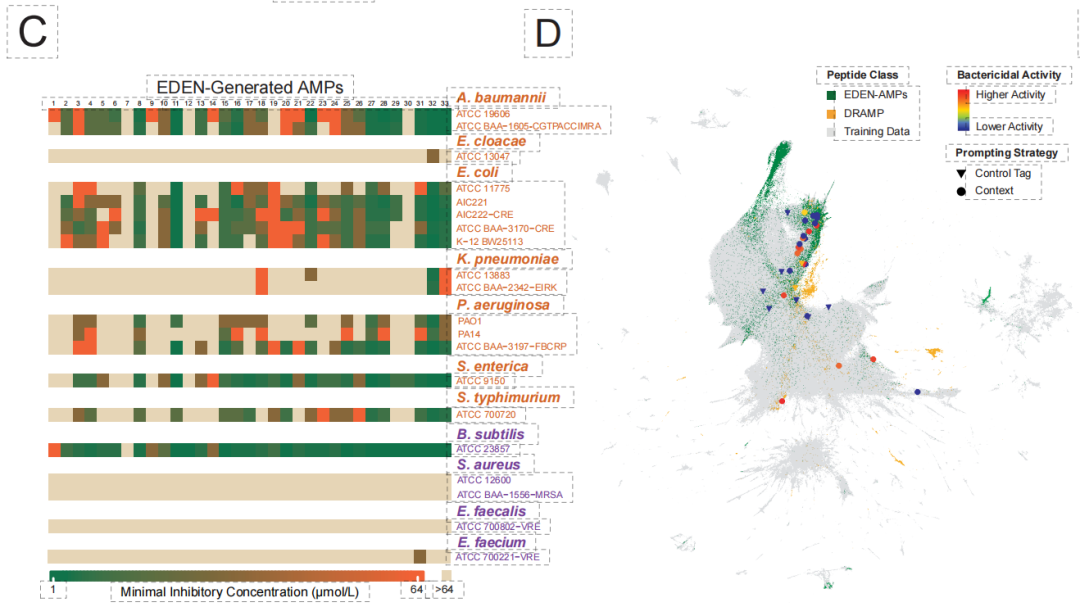
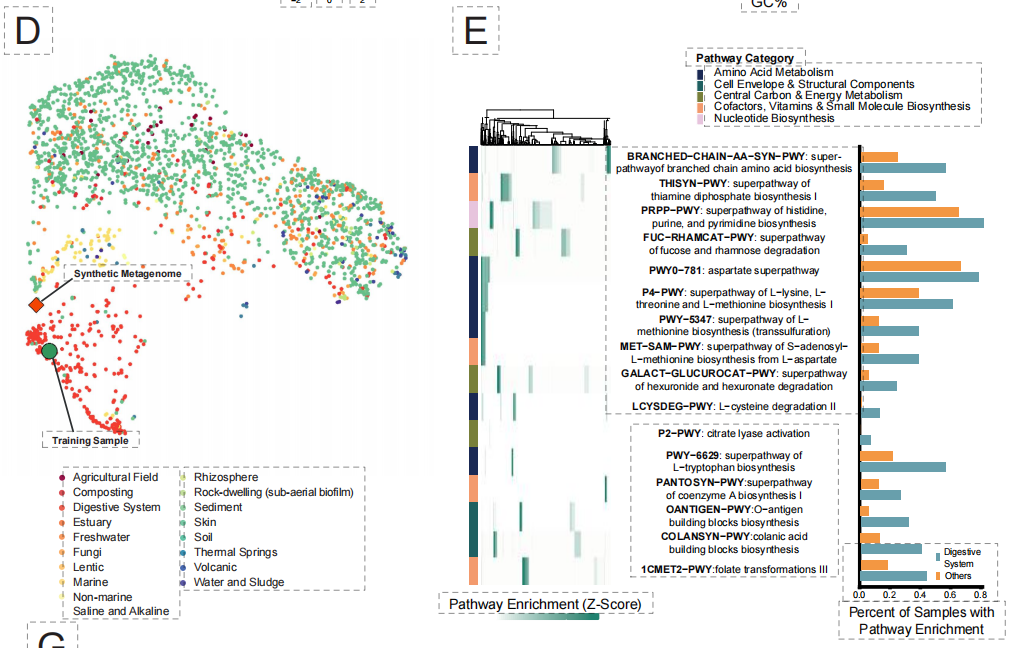
Finally, at the most complex ecosystem level, the research challenged the design of "synthetic microbiomes." Traditional methods struggle to coordinate metabolic interactions and ecological balance among multiple species. As shown in the figure below, EDEN, after fine-tuning using digestive system microbiome data,Based solely on functional genes or niche clues, a synthetic metagenomics containing over 90,000 species and spanning gigabases was successfully generated.
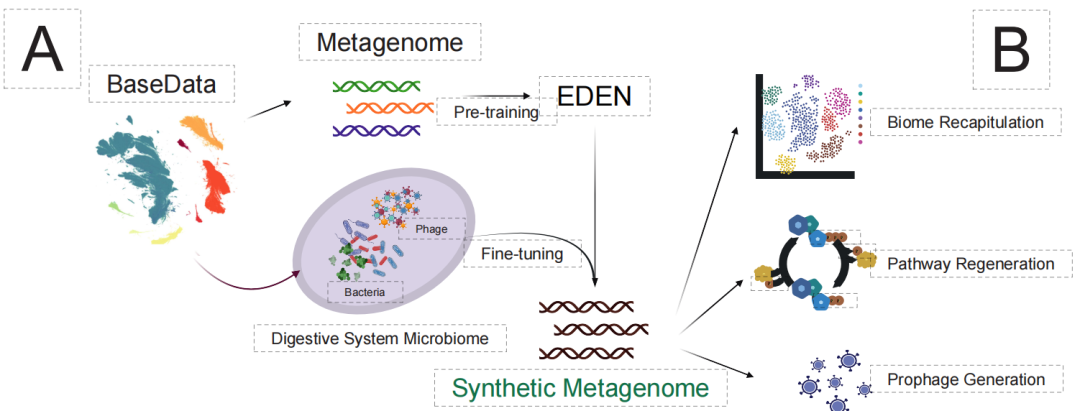
The generated results show a high degree of ecological realism:The species 99% was correctly classified into the digestive system-related biota and its cross-species metabolic pathways were fully preserved.Furthermore, the model can even accurately generate prophage structures integrated into the host genome, demonstrating that it has captured the intricate interaction logic between the host and the virus.
These four cross-scale experiments collectively demonstrate that the EDEN model, pre-trained on unified evolutionary data, can serve as a general biological design engine. It can rapidly and reliably drive therapeutic innovations from the molecular, cellular, and ecosystem levels with minimal task-specific data guidance, laying a solid practical foundation for programmable biology.
Innovation through the integration of AI and synthetic biology
In recent years, the integration and innovation of academia and industry in the field of programmable biology has accelerated significantly, and a series of major advances are redefining the boundaries of biodesign.
Leading academic institutions worldwide are transforming evolutionary wisdom into computable models with unprecedented scale and precision. For example, in early 2024, a joint team from DeepMind, Isomorphic Labs, and several universities released the AlphaFold 3 model, which can simultaneously predict protein structure, interactions, and generate novel proteins with specific functions. This model is the first to incorporate the complex interplay of biomolecules into a unified framework for high-precision simulation.Nature magazine hailed it as "a leap forward in mapping the inner workings of the molecular machines of life."
The industry is accelerating the transformation of these breakthroughs into platforms and therapies. In the field of AI-driven drug development, NVIDIA and Recursion Pharmaceuticals released BioNeMo, a biochemistry AI model library aimed at shifting drug discovery from a "needle in a haystack" to a "guided approach." Synthetic biology company Ginkgo Bioworks is leveraging its automated platform to systematically design microbial communities for carbon capture and chemical production, driving the engineering of "synthetic ecosystems."
This new wave driven by data and algorithms is propelling biology from an observational and descriptive science into a programmable, debuggable, and predictable engineering discipline. This not only means that we can more precisely code life to conquer diseases, but also foreshadows our ability to systematically design biological systems to address global challenges in the areas of resources, environment, and health.
Reference Links:
1.https://nvidianews.nvidia.com/news/nvidia-announces-broad-expansion-of-its-biomedicine-platform
2.https://www.ginkgobioworks.com/2024/01/04/ginkgo-bioworks-and-pfizer-expand-collaboration-to-advance-rna-based-therapeutics/