Command Palette
Search for a command to run...
NVIDIA Proposes ReaSyn, Which Draws on the Analogy of Thought Chain Molecular Synthesis to Achieve ultra-high Reconstruction Rate and Path diversity.
The core challenge of modern drug discovery lies in accurately identifying molecules with therapeutic potential within a nearly infinite chemical space. Traditional drug discovery often encounters a double dilemma: chemical space is incredibly vast, with the number of possible molecules consisting of just 10 atoms reaching as high as 10⁶⁰, making screening as challenging as finding a needle in a haystack. Furthermore, candidate molecules must meet multiple requirements, including activity, toxicity, and solubility. This results in development cycles often exceeding 10 years, costs in the billions of dollars, and a success rate of less than 10%.
Molecular generative models once held promise for this. Using algorithms to simulate molecular structure generation, this technology promised to significantly shorten discovery cycles and even enable "on-demand design." However, in practice, it was found that model-generated molecules were often difficult to synthesize in the laboratory. This "paper-based" limitation severely restricted their practical value.
To break through this bottleneck, the academic community has tried two strategies: one is to use "synthesizability" as the optimization goal and guide the generation of easy-to-synthesize molecules through scoring. However, due to the complexity of the structure-synthesizability relationship and the difficulty of scoring to cover experimental variables, the effect is limited; the other is to limit the model to exploring only known synthesizable molecules. Although this improves controllability, it sacrifices structural innovation. The “synthesizable projection” strategy has therefore attracted attention. Its core is to “correct” non-synthesizable molecules into analogues with similar structures and clear synthetic pathways.This strategy can flexibly integrate multiple generation methods to support tasks such as active compound expansion and lead optimization.
In this context,ReaSyn, an efficient and synthesizable molecular projection framework with integrated reasoning capabilities, launched by the NVIDIA research team,By adopting the Chain-of-Reaction (CoR) representation and considering the synthesis path as the Chain-of-Thought (CoT) reasoning path of LLM, a new path has been opened up for solving the practical problems of molecular synthesis.
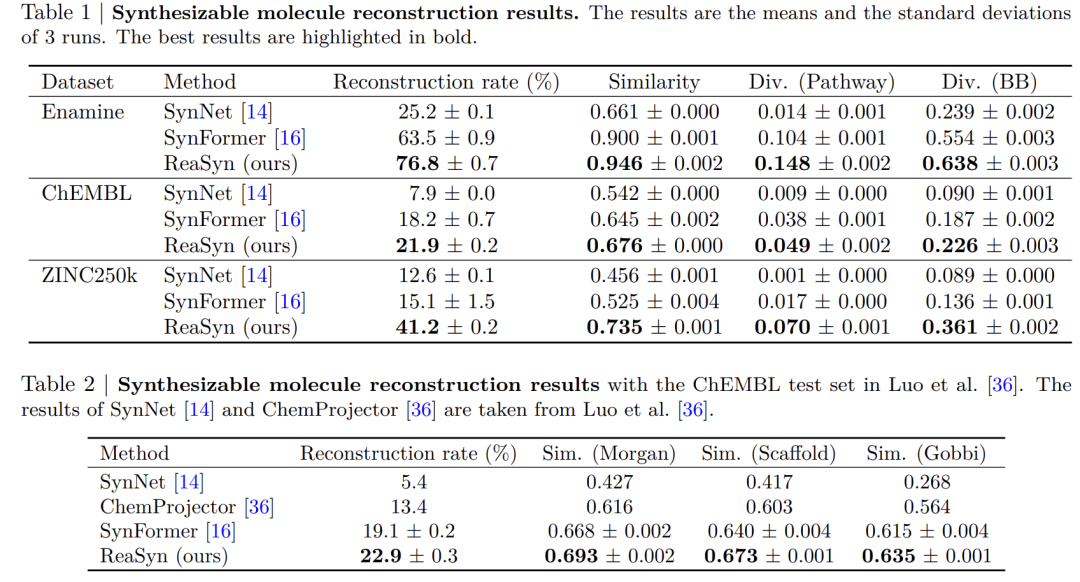
In the reconstruction of synthesizable molecules,ReaSyn achieved the highest reconstruction rate and path diversity;It also achieved the best optimization performance in synthesizable target-directed molecular optimization and significantly outperformed previous methods in the synthesizable hit expansion task.
The relevant research results were published on arXiv under the title "Rethinking Molecule Synthesizability with Chain-of-Reaction".
Research highlights:
* This study proposes the ReaSyn framework and chain of reaction (CoR) representation to transform synthetic pathways into explainable thought chains for reasoning.
* Customized RL fine-tuning and computational expansion solutions significantly improve the model's exploration efficiency and optimization performance.
* Through multi-task experiments, the effectiveness and versatility of the framework in the generation and optimization of synthesizable molecules are confirmed.
Paper address:
https://arxiv.org/abs/2509.16084
Follow the official account and reply "ReaSyn" to get the complete PDF

More AI frontier papers:
Construction of a dataset close to real drug development
The study first constructed an experimental framework close to the real drug development scenario, using a reaction set containing 115 common reaction types and combining it with 212,000 purchasable building blocks obtained from the Enamine US inventory catalog.Together, they define a synthetic chemical space exceeding 10⁶⁰ molecules in size.The experiment focuses on the task of "reconstructing synthesizable molecules", aiming to test the model's ability to cover a large chemical space by generating feasible synthetic pathways for given molecules.
In the test set design, the research team used multiple sets of molecules with different challenges.In addition to a baseline test set of 1,000 molecules randomly selected from the Enamine REAL diversity dataset and the ChEMBL database, an extended test set was constructed to simulate the real-world scenario of "building block inventory updates" in drug development. Over 37,000 molecules with fewer than 18 heavy atoms were selected from the ZINC250k library as new building blocks, and 1,000 test molecules were generated from this extended inventory. The experiment also incorporated the ChEMBL test set proposed by Luo et al. to ensure comparability with existing research.
ReaSyn Framework: A Progressive Technology Route from Molecular Representation to Inference Enhancement
The ReaSyn framework aims to address key inference bottlenecks in the projection of synthesizable molecules.Its technical route follows a progressive logic from innovation in molecular representation to enhanced reasoning capabilities.
As shown in the figure below, the study first clearly defined the synthesizable chemical space: this space is determined by a set of building blocks and a set of reaction rules. Each reaction describes the transformation from reactants to products using the SMARTS language, while the synthesizable space represents the set of all products that can be obtained from the initial building blocks through the iterative application of reaction rules. Within this framework, the core goal of the synthesizable projection is to generate a synthetic pathway 𝑝 for a given target molecule 𝑥 such that the structural similarity between the end product of the pathway and 𝑥 is maximized.
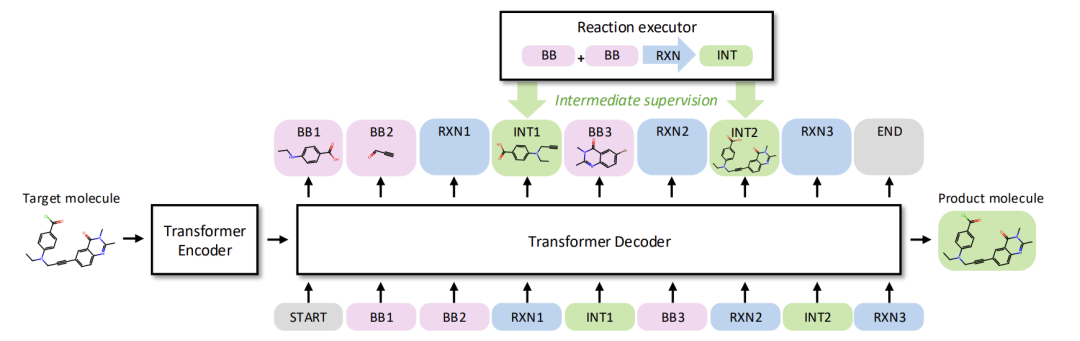
ReaSyn's overall framework
In terms of the representation of the molecular synthesis pathway, as shown in the figure below,ReaSyn innovatively proposed the "Chain-of-Reaction (CoR)" representation strategy.This overcomes the various limitations of the traditional "synthetic tree suffix representation." While traditional methods support autoregressive generation, they suffer from inherent flaws such as the need for implicit learning of reaction rules, the susceptibility of hierarchical predictions to error propagation, and the lack of bijectivity in building block fingerprint representations.
The reaction chain representation method has achieved three important breakthroughs while maintaining its versatility: integrating the chain of thought (CoT) at the chemical reaction level, achieving complete path prediction without hierarchical classification, and eliminating dependence on molecular fingerprints.
In specific implementation, the synthesis pathway is decomposed into multiple functional blocks, all of which share a unified vocabulary. Molecular blocks are represented by SMILES strings with special tags, while reaction blocks are represented by single tokens. Finally, a complete pathway sequence is formed through splicing operations.
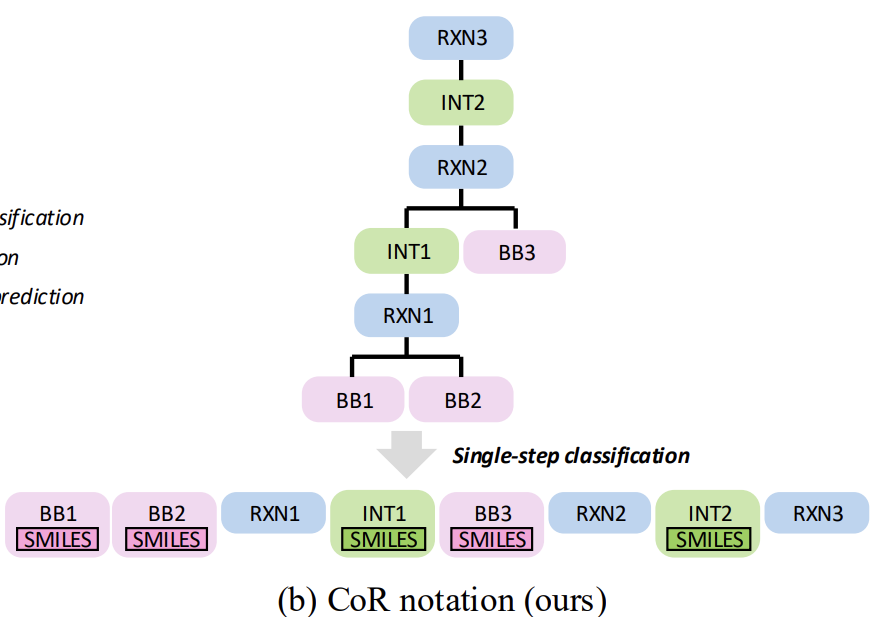
The model training adopts a two-stage strategy that combines supervised learning and reinforcement learning finetuning.
In the supervised learning phase, the paired data of target molecules and synthetic pathways are used to train the Transformer model with the goal of predicting the next token.And by designing a token type weighted loss function to balance the learning intensity of different types of tokens, while providing richer supervision signals with the help of intermediate products.The online reinforcement learning algorithm is used in the reinforcement learning fine-tuning stage.The reward mechanism is used to guide the model to explore more effective paths. Its loss function not only considers the maximization of path rewards, but also focuses on the stability of model behavior, effectively making up for the limitations of supervised learning in exploration capabilities.
In the inference stage,ReaSyn combines a stack structure with a beam search mechanism to achieve goal-directed test-time compute scaling and customize scoring strategies based on different task requirements. The stack dynamically manages reactants and intermediates, supporting the step-by-step reasoning process; beam search maintains search diversity by maintaining multiple high-scoring candidate paths.
In the molecular reconstruction task,The scoring strategy focuses on structural similarity and reaction feasibility to ensure accurate reproduction of the target molecule; in the molecular optimization and activity expansion tasks, a reward model is introduced to evaluate the target properties of building blocks and intermediates, guiding the search towards synthesizable molecules with ideal properties, and realizing targeted exploration and optimization within the synthesizable space.
Experimental results: Multi-task performance surpasses SynNet and other methods, and ablation experiments verify the effectiveness of core components
The experimental results are shown in the following table.ReaSyn demonstrates superior comprehensive performance over existing SynNet and SynFormer methods in multiple key tasks.
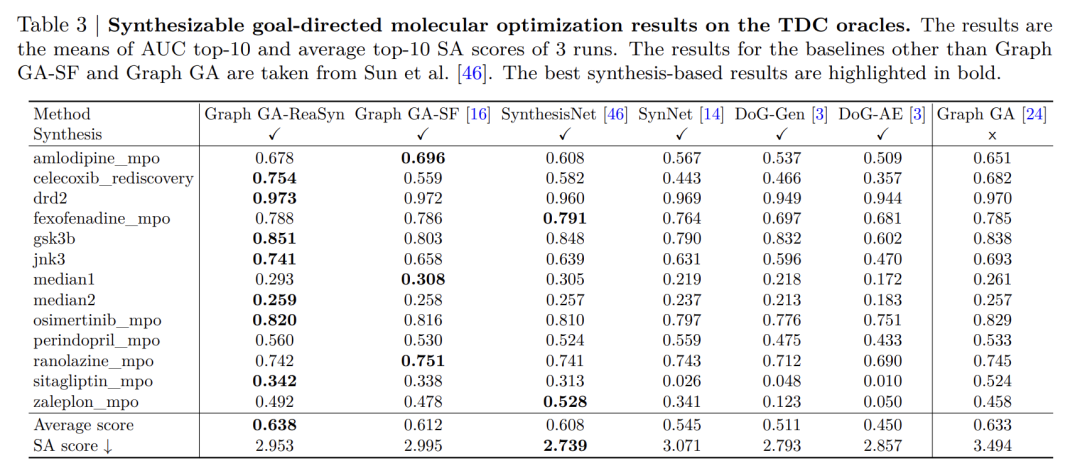
In the task of optimizing synthesizable target molecules,The research focused on how ReaSyn can improve the practicality of traditional optimization methods.This task uses the graph genetic algorithm (Graph GA) as the basic framework, and introduces ReaSyn after its breeding step to perform synthesizable projection processing to ensure that the obtained molecules are all in the synthesizable space. This method is named Graph GA-ReaSyn.
The experiment is divided into two parts: one is the general property optimization based on the TDC oracle function, and the other is the targeted optimization of the binding affinity for soluble epoxide hydrolase (sEH).
In the TDC mission,As shown in the table below, Graph GA-ReaSyn outperformed all synthetic constraint-based baseline methods in the "AUC top-10" metric across 15 optimization tasks, and its synthetic accessibility score (SA score) was significantly better than the original Graph GA, indicating that ReaSyn effectively improved synthesizability while maintaining optimization performance. In sEH affinity optimization, ReaSyn outperformed methods such as FragGFN, SynFlowNet, and SyntheMol in terms of binding affinity, SA score, and drug-likeness (QED).In particular, its advantages in balancing target properties and synthesizability are highlighted.
In the task of expanding the range of synthetic active compounds,ReaSyn uses beam search to generate multiple structurally similar and synthesizable analogs of known active compounds to expand the candidate molecule library. The experiment used JNK3 inhibitors as the research target, selecting the top ten rated molecules from the ZINC250k database as a starting point and generating 100 analogs for each molecule.Evaluated by three indicators, namely, “analog rate,” “improvement rate,” and “success rate,” ReaSyn outperformed previous methods in all indicators.
AI-driven synthetic pathway prediction promotes innovation in the design of synthesizable molecules
While AI-driven synthesis pathway prediction technologies such as ReaSyn are developing, global academia and business communities are also actively exploring this field, driving innovation in the design of synthesizable molecules from different paths.
Academic research often focuses on breakthroughs in new methods and underlying mechanisms. For example,Organa, a desktop robotic system developed by the University of Toronto,By combining computer vision and a large language model (LLM), it is possible to convert natural language instructions into standard chemical description language χDL code, automate some chemical laboratory tasks, and thus convert scientists' verbal instructions into experimental processes.
Mobile Robotic Chemist, an AI chemist independently developed by the University of Liverpool,688 experiments were completed in 8 days, 1,000 catalytic formulas were studied in a week, and a new catalyst was discovered.
Innovation in the business world focuses more on transforming advanced technologies into actual productivity and integrating them into existing workflows.The strategic cooperation between BenevolentAI, a British company specializing in artificial intelligence drug research and development, and Merck is very representative.The former relies on the chemical design tools of its end-to-end AI platform, combined with wet laboratory facilities in Cambridge, UK, to provide Merck's drug research and development pipeline with full-chain support from active compound identification to preclinical candidate molecule development. The core lies in utilizing the synthetic pathway reasoning capabilities of large language models to ensure that the generated small molecule compounds have both high activity and synthesizability, significantly shortening the conversion cycle from concept to candidate molecules.
Insilico Medicine, an AI-driven biotech company, has demonstrated the practical value of synthetic accessibility design in end-to-end drug development. Its idiopathic pulmonary fibrosis candidate drug INS018_055, designed using generative AI, uses the built-in ReaSyn-like synthesizable projection module toAchieved a 100% success rate in synthesizing TP3T in preclinical studies,Moreover, it only takes 18 months from target discovery to candidate molecule determination, which is 60% shorter than the industry average.
These diverse explorations from academia and industry, although with different entry points and technical paths, all point to one goal: to improve our ability and efficiency in designing and synthesizing useful molecules through innovative methods, and ultimately inject new impetus into many fields such as drug research and development and new material development.
Reference Links:
1.https://mp.weixin.qq.com/s/Mz64afMOOI_7m-Nqg_m5oQ
2.https://mp.weixin.qq.com/s/1Juv9z1-mUOR6Sip4KwvgQ
3.https://mp.weixin.qq.com/s/vhhb2OUtCRpbPLg8j4YsYQ