Command Palette
Search for a command to run...
One-stop Protein zero-shot Mutation prediction/function Prediction, Protein Engineering Workbench VenusFactory Enables full-stack Development
AI's significant improvement in design efficiency is reshaping the traditional research paradigm in protein design. Compared to traditional protein design experiments, large AI models can not only predict and screen protein sequences, structures, and functions in a fraction of the time, but can also design novel proteins not found in nature based on physical and chemical principles and data patterns. They can even simultaneously predict protein properties such as stability, binding affinity, and kinetics through multi-task learning and deep learning models.
However,The complex computational framework of the model and the huge protein database have raised the threshold for using AI tools.On the one hand, the protein design field's reliance on biological data requires researchers to retrieve, download, compile, and convert data from multiple databases, resulting in a significant amount of time-consuming. On the other hand, protein AI models can currently only solve individual tasks in niche areas and lack an evaluation system with authoritative benchmark data.
In addition, regarding the challenges of AI protein design, Dr. Tan Yang from Professor Hong Liang's research group at Shanghai Jiao Tong University also introduced that the existing AI models in the field of protein design not only have difficulty in acquiring data and unifying the format, but also have difficulty in adjusting parameters and slow training speed.The obstacles caused by "data barriers, model barriers, and application barriers" have hindered the popularization and application of AI tools in a wider scientific research community.
At the same time, as far as existing solutions are concerned, web servers are simple and easy to use, but they have limited functions, cannot be trained based on individual data, and have limited intelligence. Agents reduce human intervention, can directly focus on the result goals, and autonomously complete one or more work units.
To promote the widespread application of artificial intelligence in the field of protein engineering,Professor Hong Liang's research group at Shanghai Jiao Tong University developed a one-stop open-source protein engineering workbench, VenusFactory.The platform integrates biological data retrieval, standardized task benchmarking, and pre-trained protein language models (PLMs). The platform combines the dual functions of a web server and an agent:
* Implemented 0-code customization of AI models using private datasets, supporting command line execution and Gradio-based codeless interface.
* Provides open-source downloads of 30+ large model evaluation benchmark datasets, integrates more than 40 protein-related datasets and more than 40 popular PLMs, and easily links to protein data.
* It can achieve zero-sample mutation prediction, automatically combine AI models to recommend mutations based on demand, and incorporate supervised prediction modules to predict properties through target integration AI models.
at present,The VenusFactory protein engineering design platform is now available in the tutorial section of HyperAI's official website (hyper.ai). The VenusFactory platform tutorial covers 7 functional modules, and you can experience it online with one-click deployment:
* Training: Zero-code model training, supports 40+ large models, and uses private datasets to train your own models.
* Evaluation: An easy-to-use tool for comprehensive performance evaluation of protein models.
* Prediction: Use the trained model to predict the function of new protein sequences.
* Quick Tools: Easy-to-use version, supporting zero-sample mutation prediction (directed evolution) and supervised prediction (function or property prediction).
* Advanced Tools: Advanced customized version, supporting zero-sample mutation prediction (directed evolution) and supervised prediction (function or property prediction).
* Download: Easily link to protein data and support multi-threaded downloading of major mainstream databases (RCSB, UniProt...).
* VenusAgent: A protein engineering agent that works with DeepSeek to enable AI protein computation.
Tutorial Link:
In addition, we have prepared surprise computing resource benefits for new users.Register with the invitation code "VenusFactory" to get 2 hours of dual-SIM A6000 usage time (resource validity period is 1 month).The quantity is limited, don’t miss it!
Demo Run
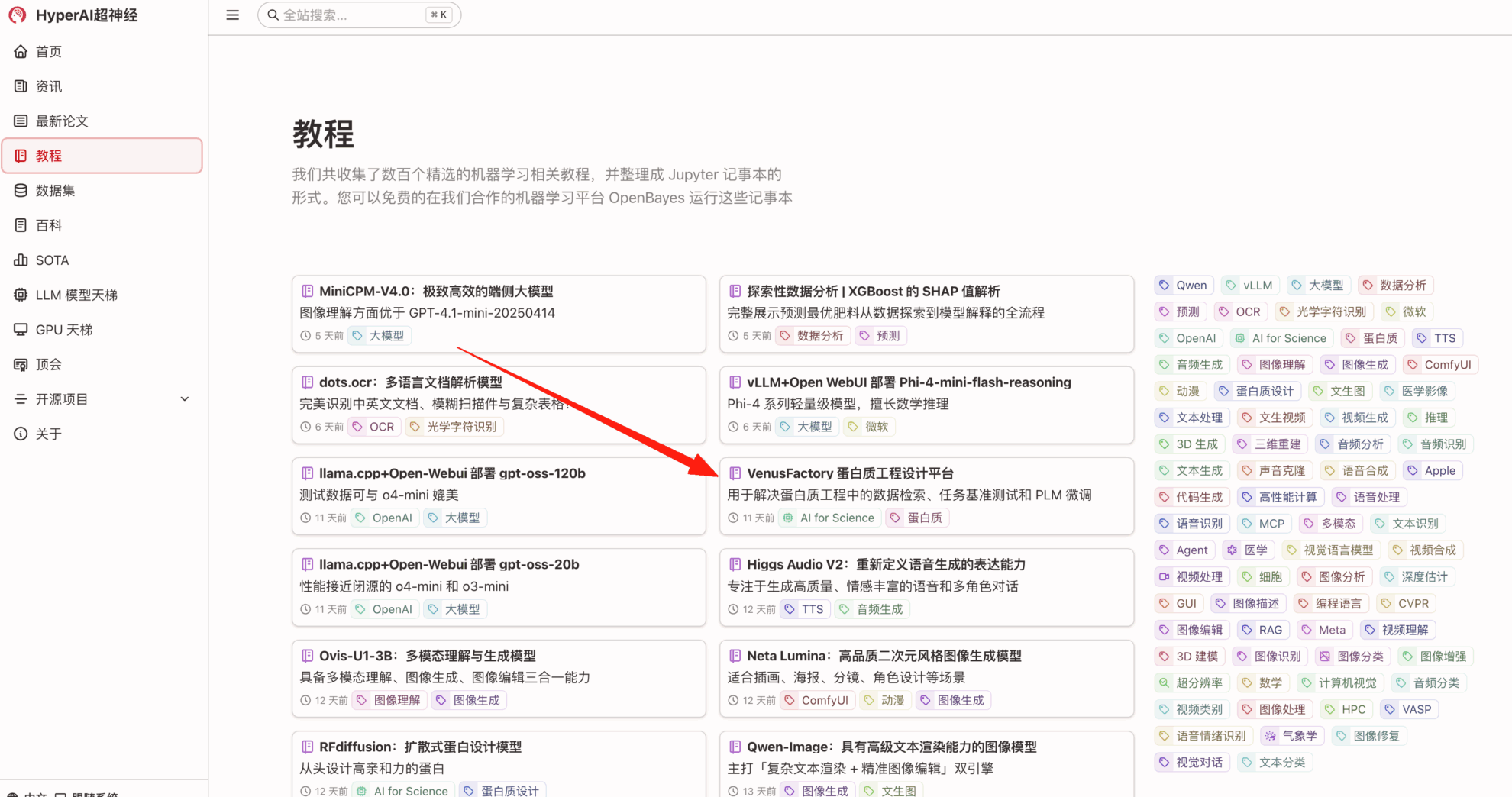
1. Enter the URL hyper.ai in your browser. Once you reach the homepage, click the Tutorials page, select VenusFactory Protein Engineering Platform, and click Run this tutorial online.
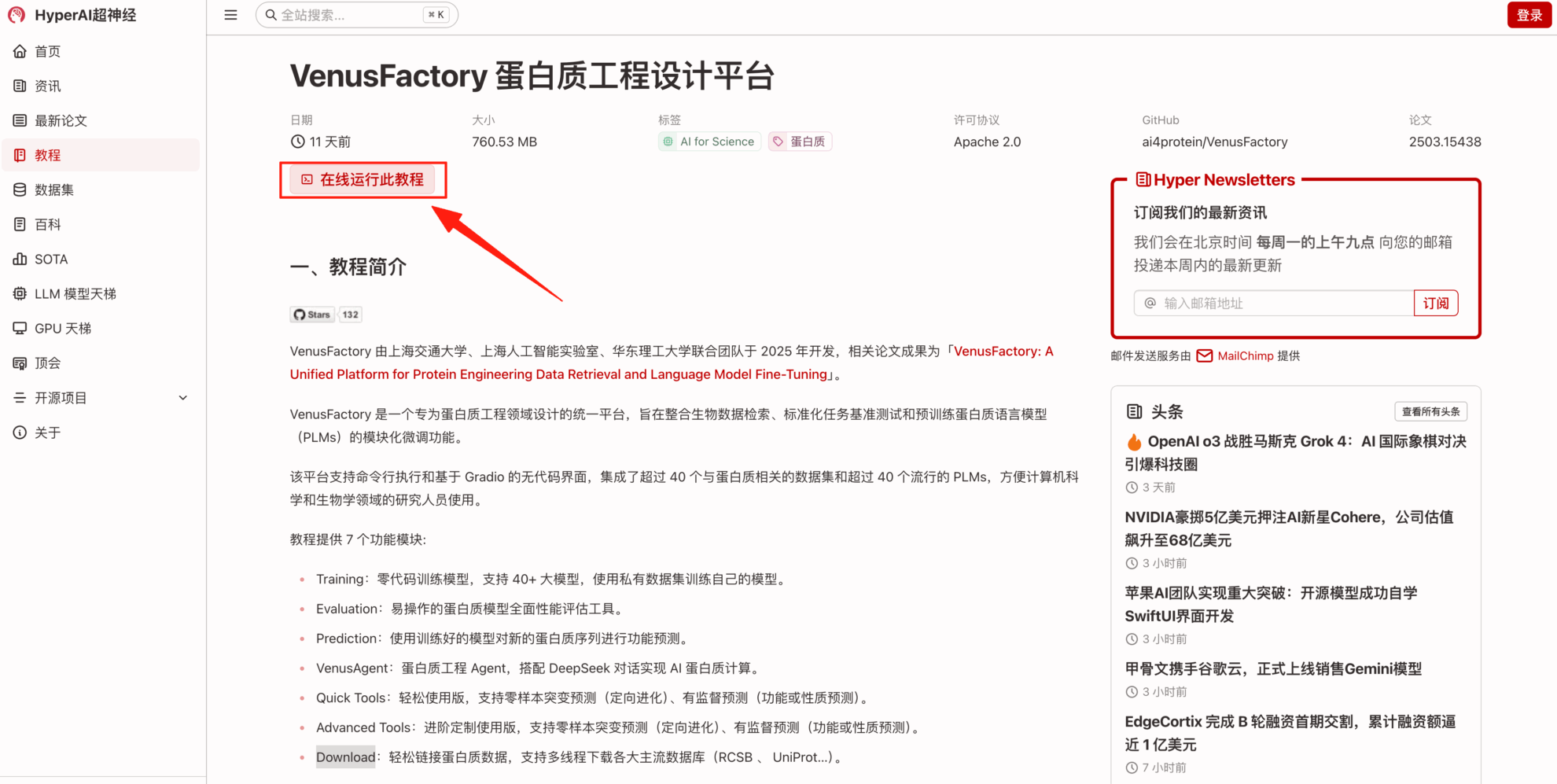
2. After the page jumps, click "Clone" in the upper right corner to clone the tutorial into your own container.
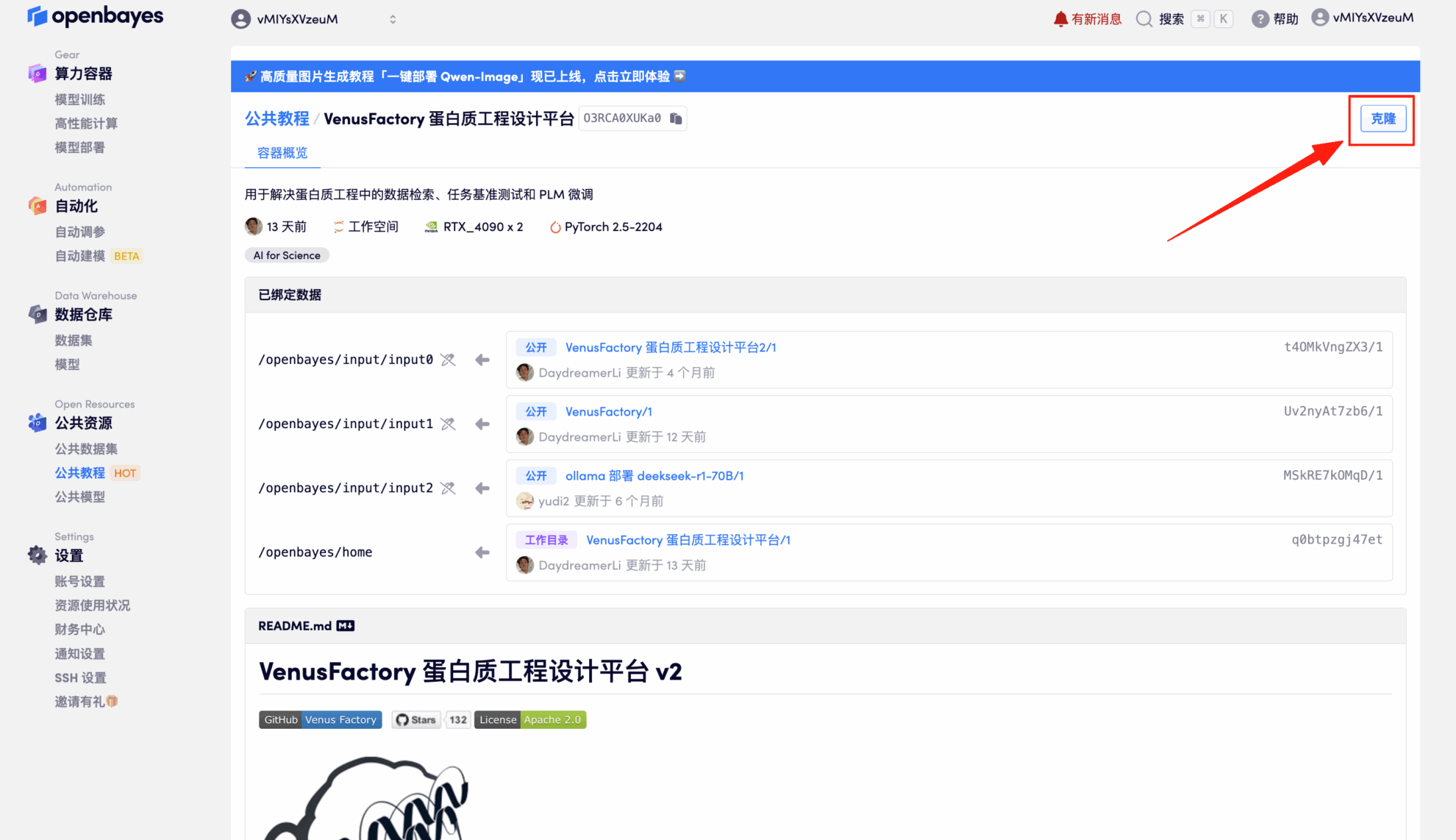
3. Select the NVIDIA GeForce RTX 4090-2 and PyTorch images and click Continue. The OpenBayes platform offers four billing options: pay-as-you-go or daily/weekly/monthly plans. New users can register using the invitation link below to receive 4 hours of free RTX 4090 and 5 hours of free CPU time!
HyperAI exclusive invitation link (copy and open in browser):
https://openbayes.com/console/signup?r=Ada0322_NR0n
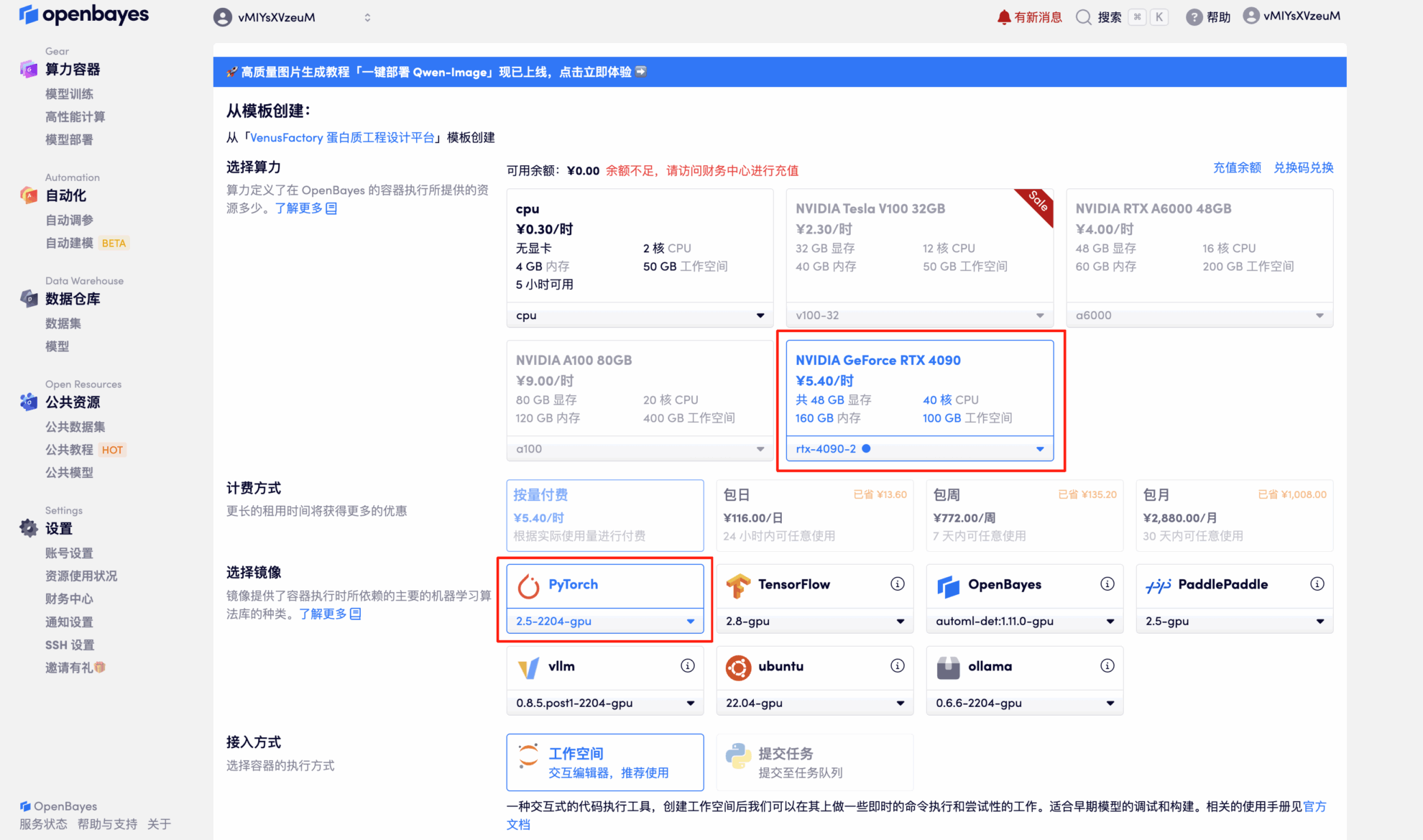
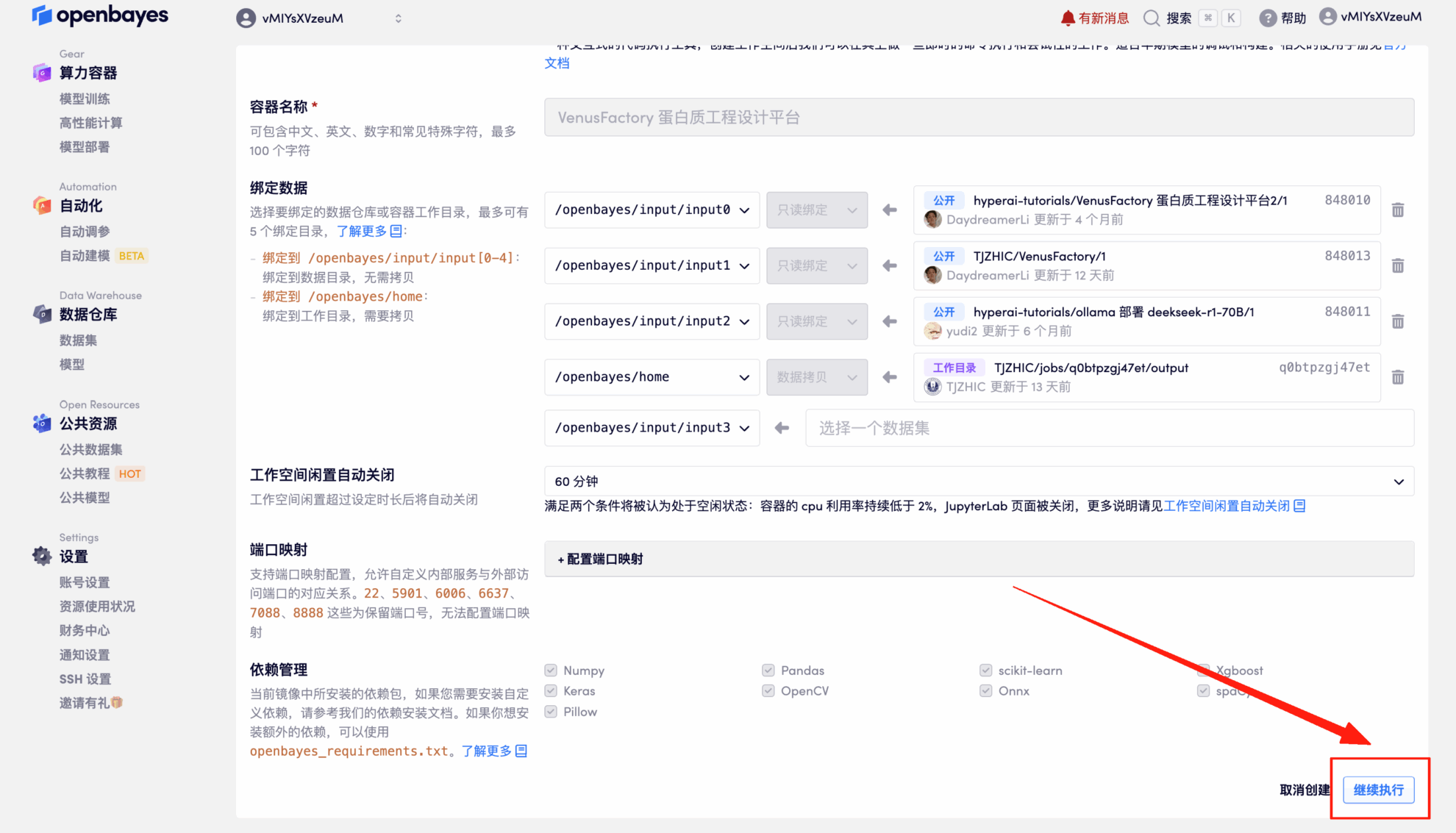
4. Wait for resources to be allocated. The first cloning process will take about 2 minutes. When the status changes to "Running," click the arrow next to "API Address" to jump to the Demo page.Since the model is large, it takes about 3 minutes for the WebUI interface to be displayed, otherwise "Bad Gateway" will be displayed.Please note that users must complete real-name authentication before using the API address access function.
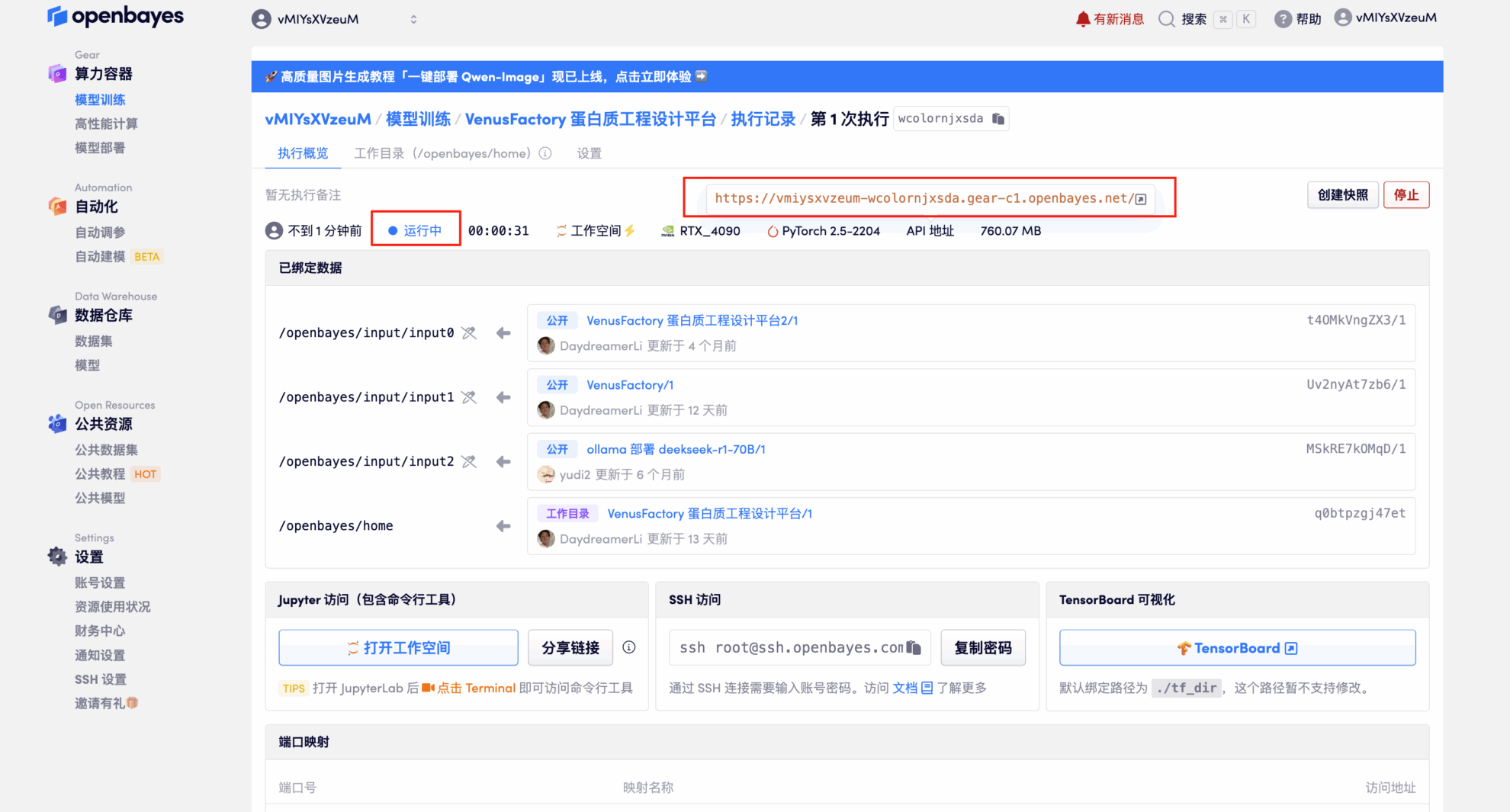
Effect Demonstration
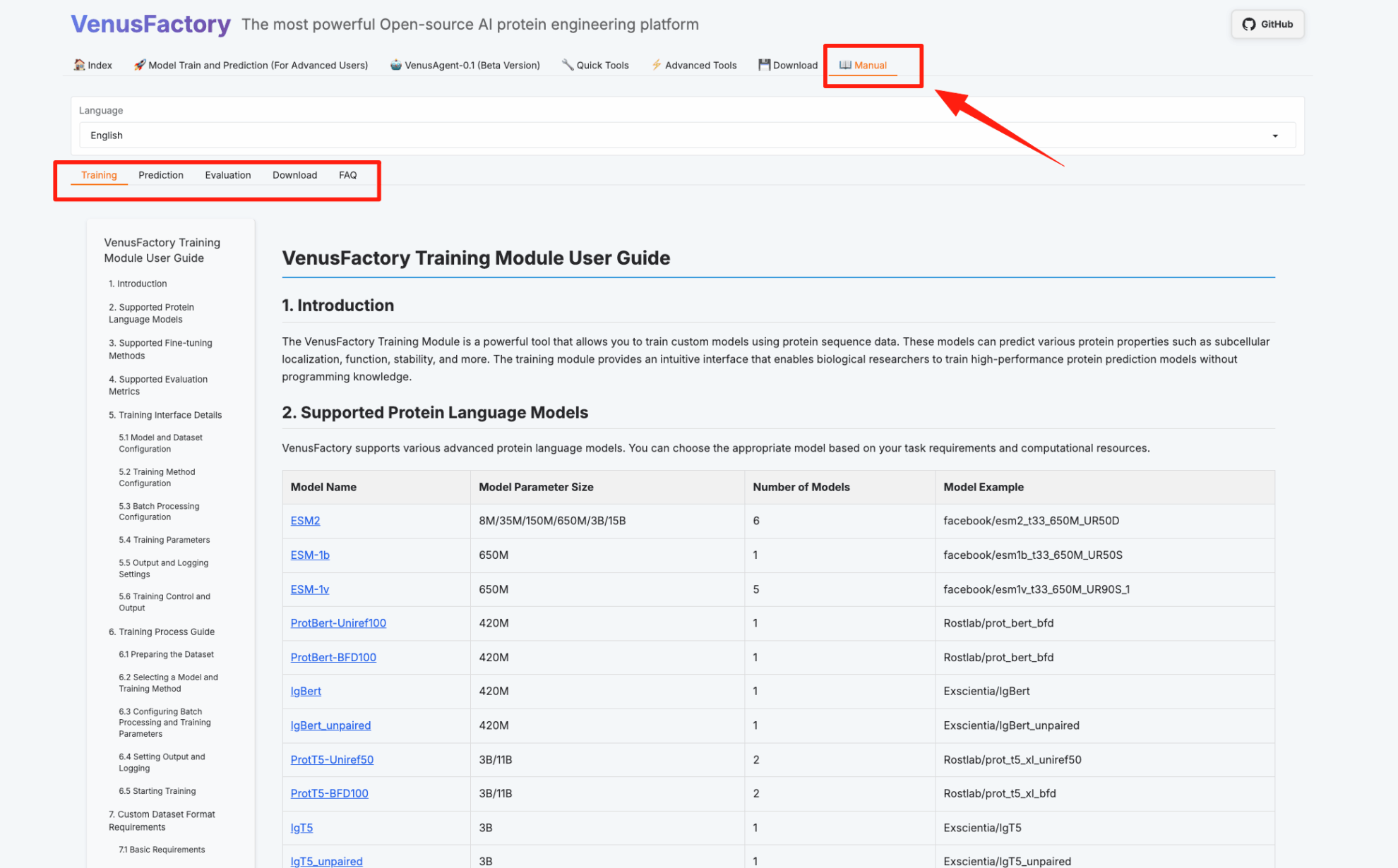
The following is the VenusFactory usage page. Click "Manual" to directly view the usage guides of the training module, prediction module, evaluation module, and download module:
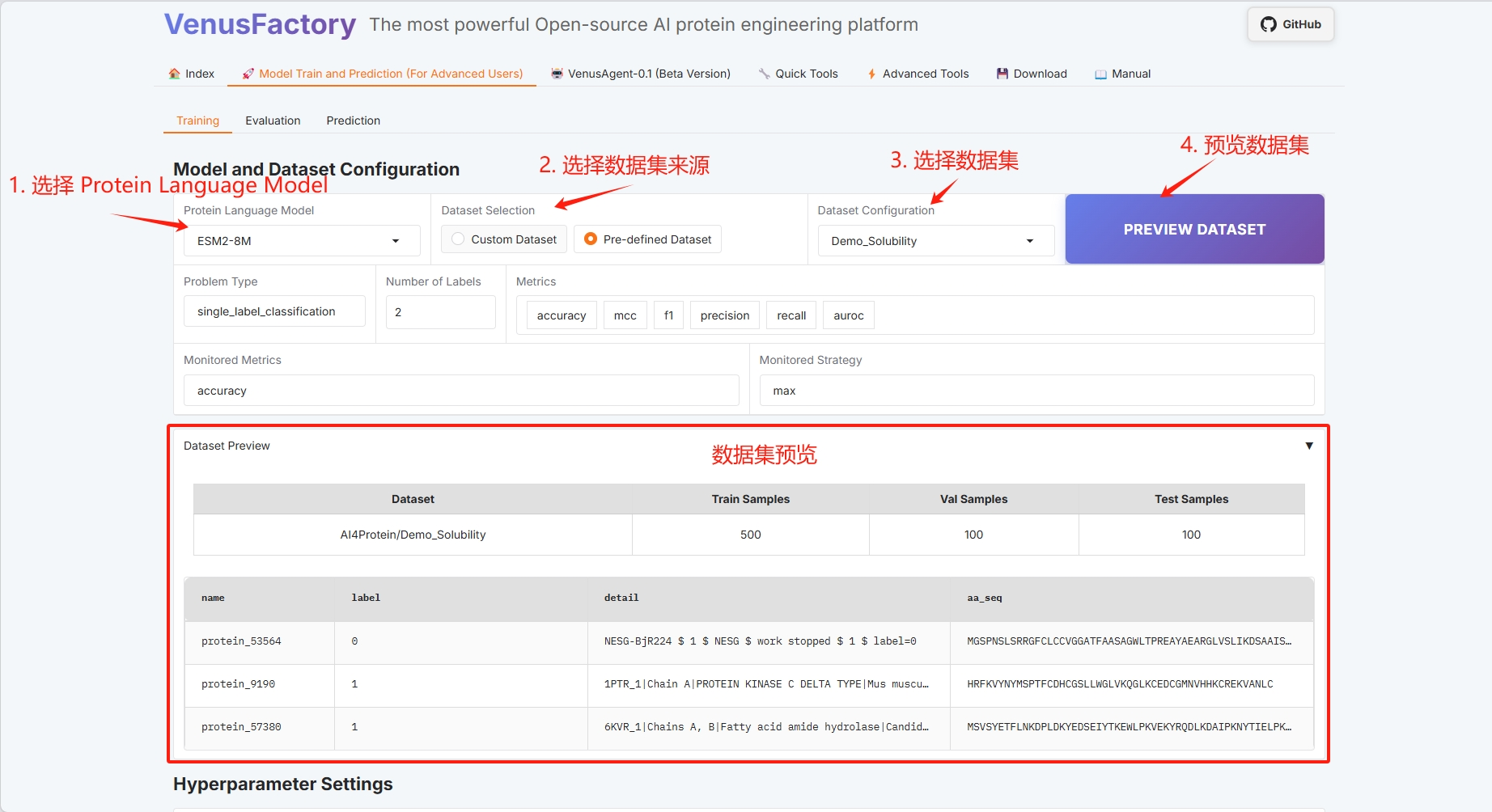
Training module display
Click the “Training” module in the “Model Train and Prediction Training” module:
* Select Protein Language Model
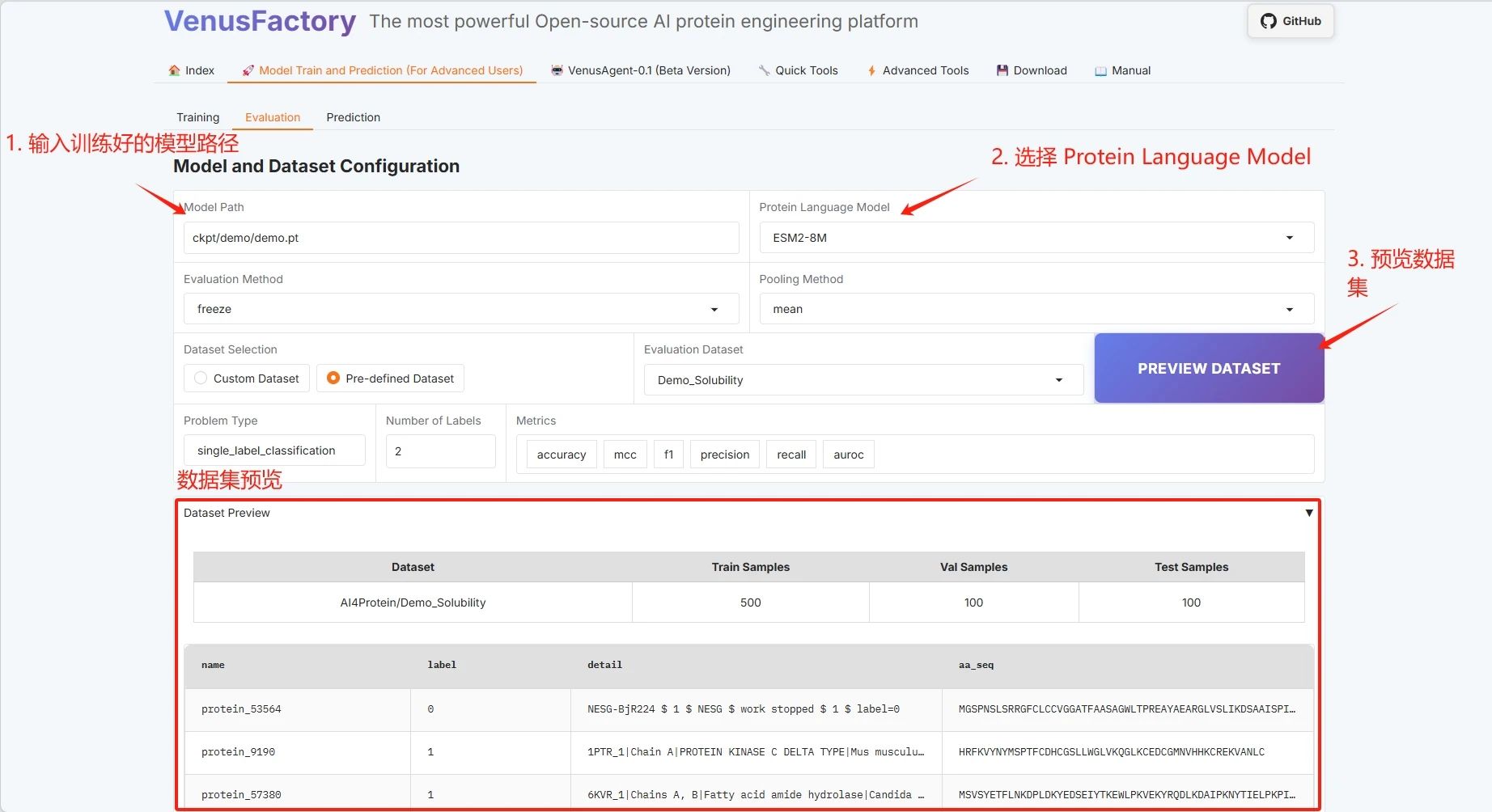
* Dataset selection
* Dataset preview
* Training method configuration (refer to the user guide for specific information)
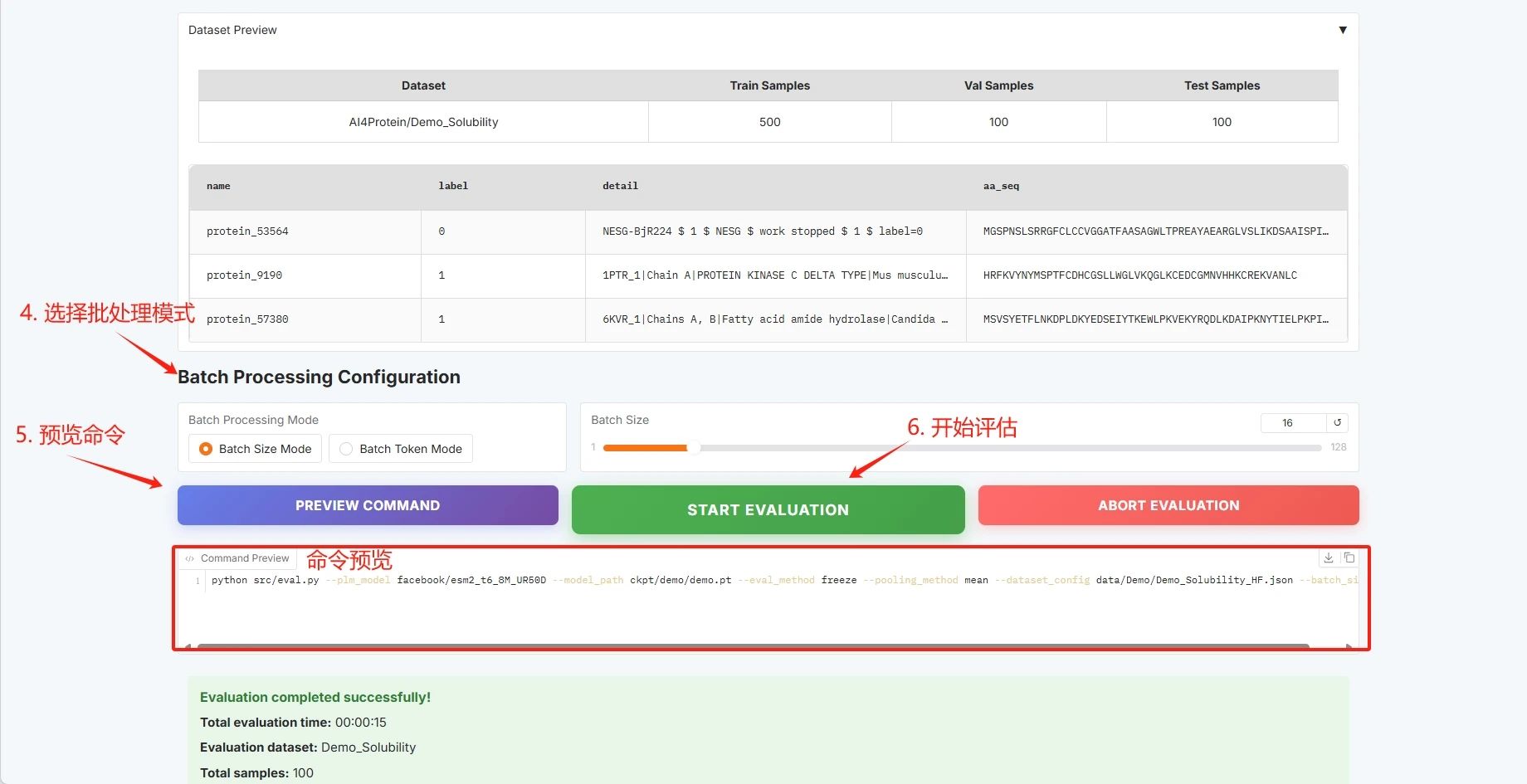
* Batch configuration (see the User Guide for details)
Set the training model save path and click "START TRAINING" to start training.
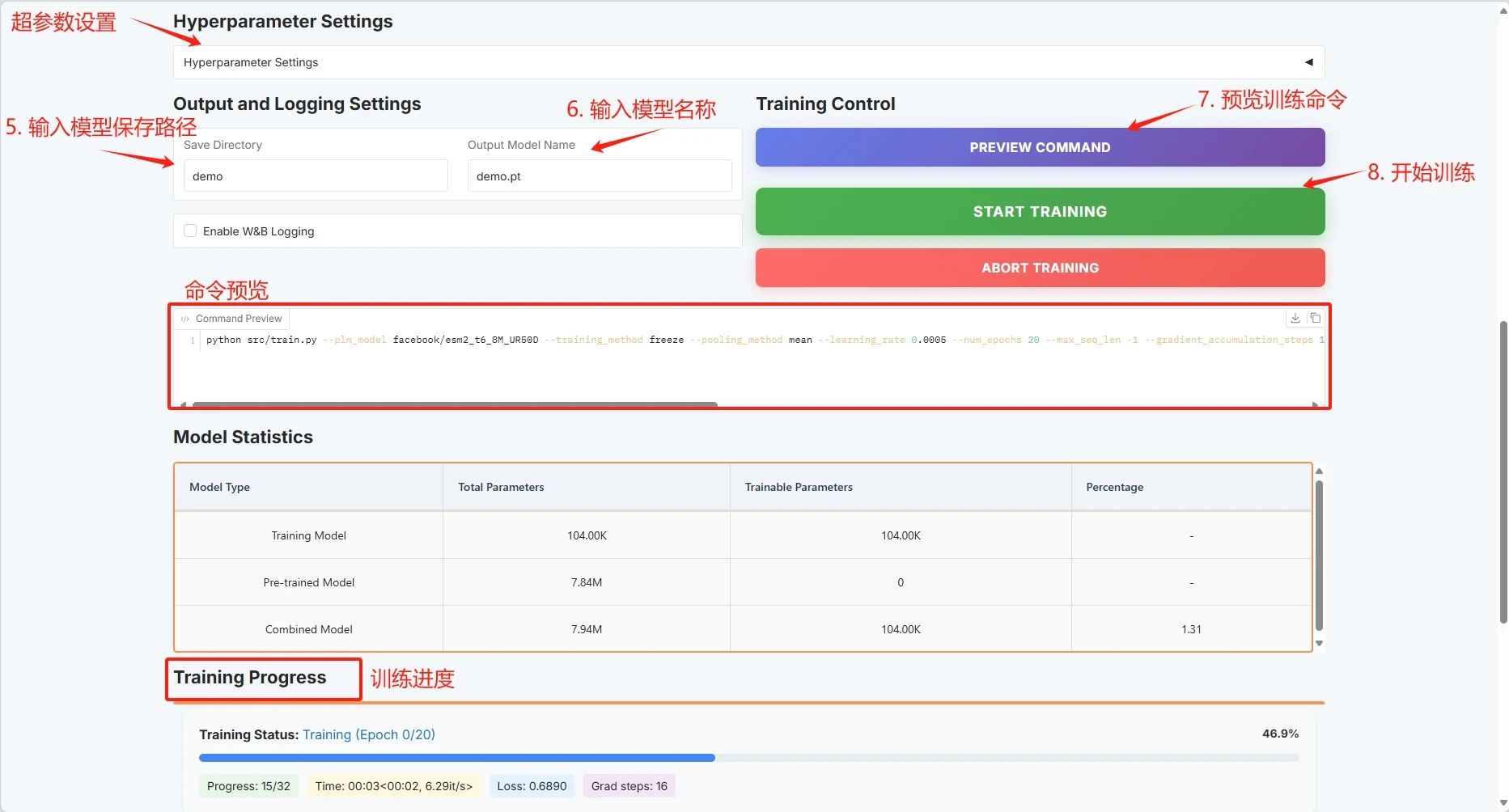
At this point you can see the training parameters and loss curve:
If you want to use your own dataset, you can use the Custom Dataset configuration. Just fill in the path of your dataset (see the Manual documentation for details).
Evaluation module display
Click the “Evaluation” module in the “Model Train and Prediction Training” module.
Batch configuration, click "START EVALUATION" to start training.
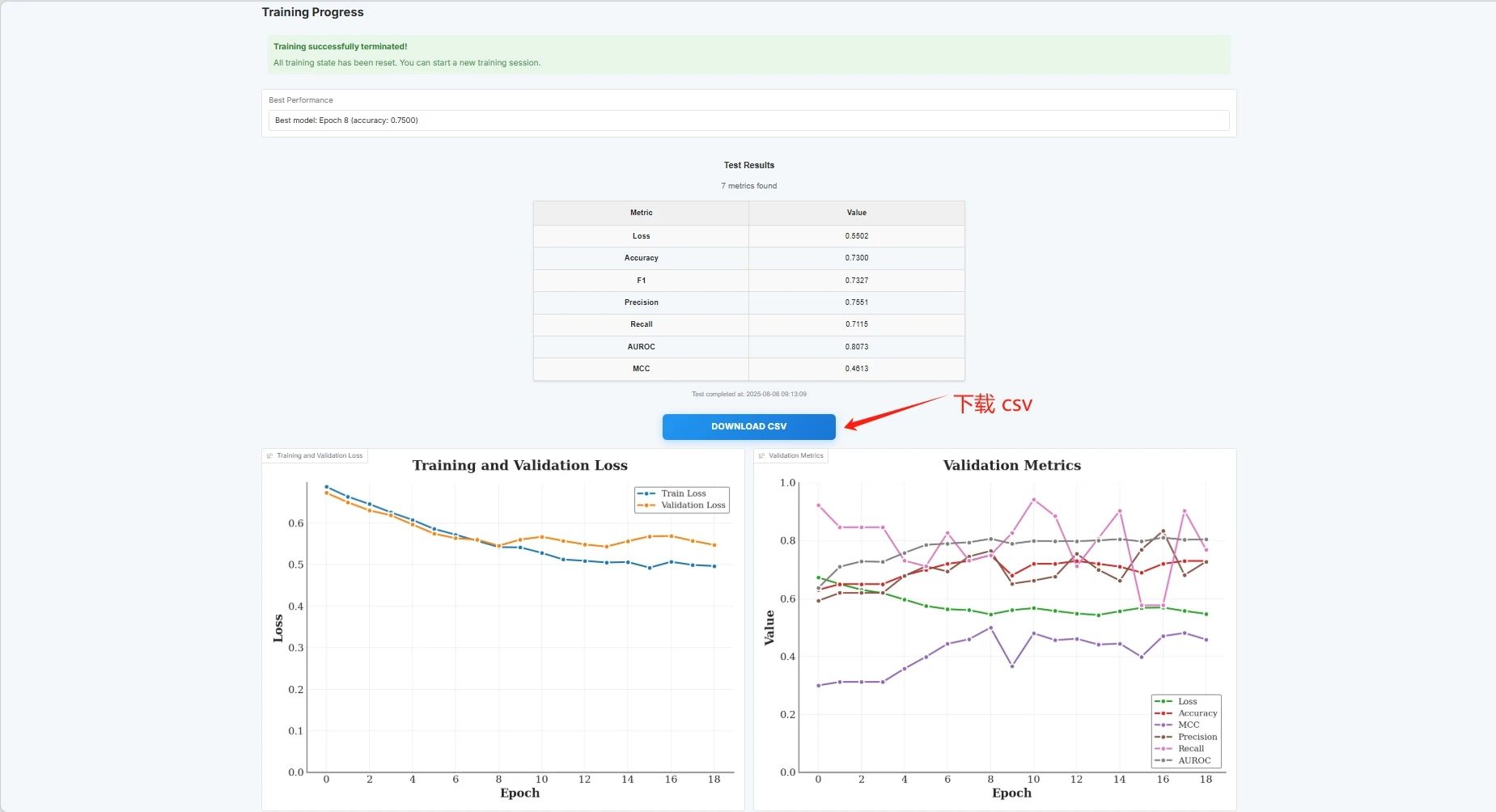
The evaluation results are as follows, and you can download the CSV file:
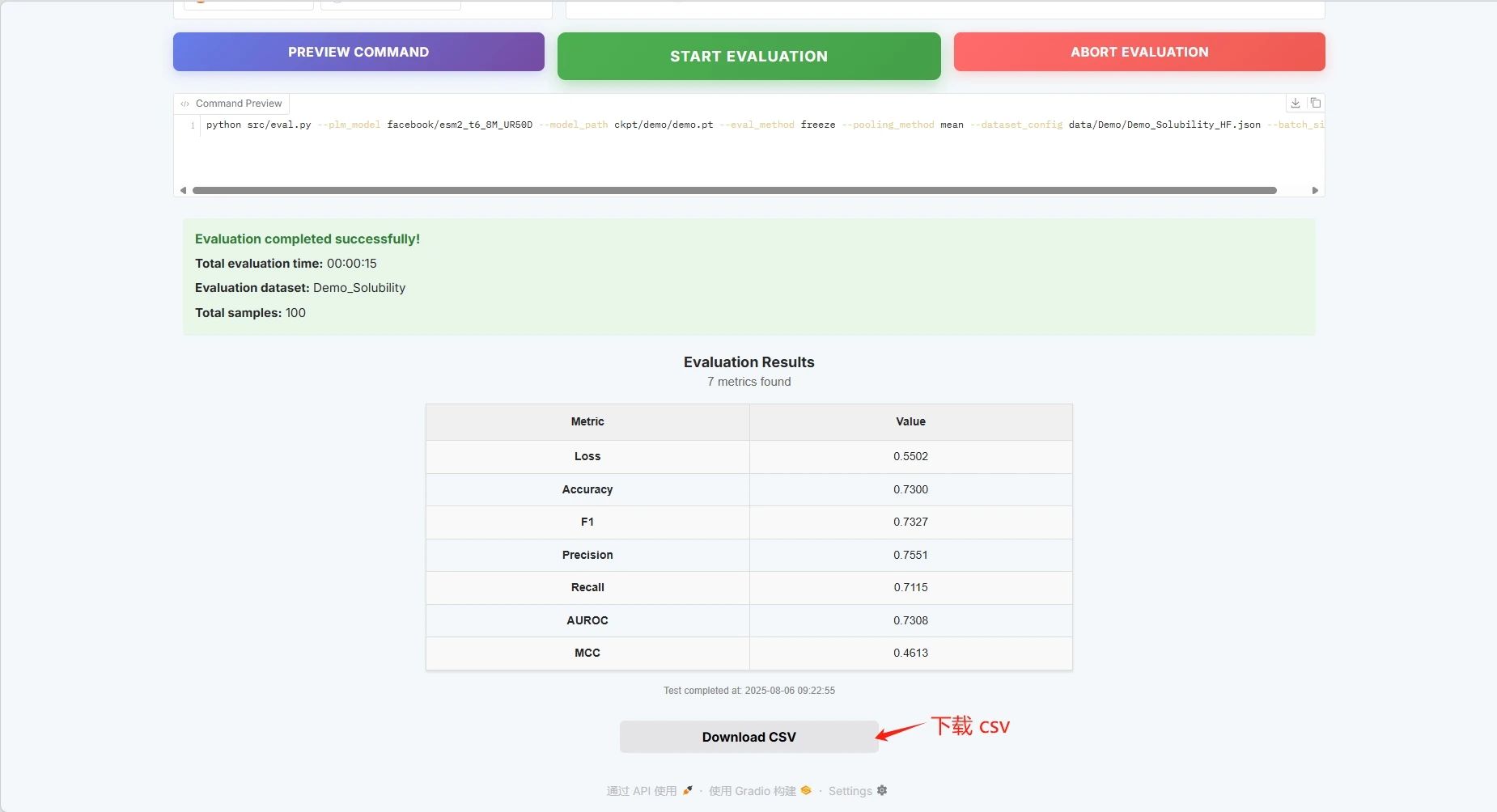
If you want to use your own dataset, you can use the Custom Dataset configuration. Just fill in the path of your dataset (see the Manual documentation for details).
Prediction module display
Click the "Prediction" module in the "Model Train and Prediction Training" module, set the training model save path, select the protein language model, and click "START PREDICTION" to start training.
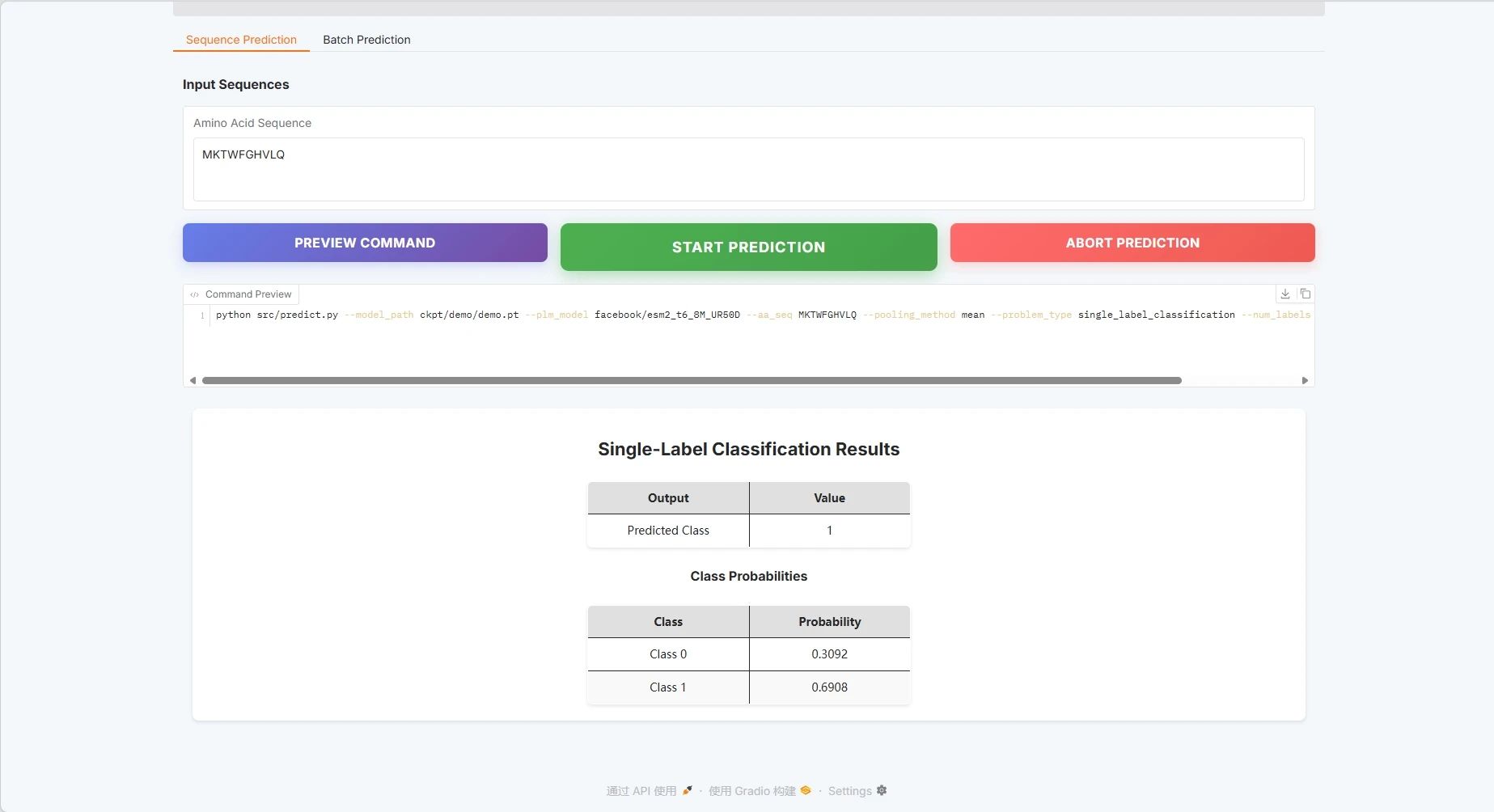
Take single sequence prediction as an example:
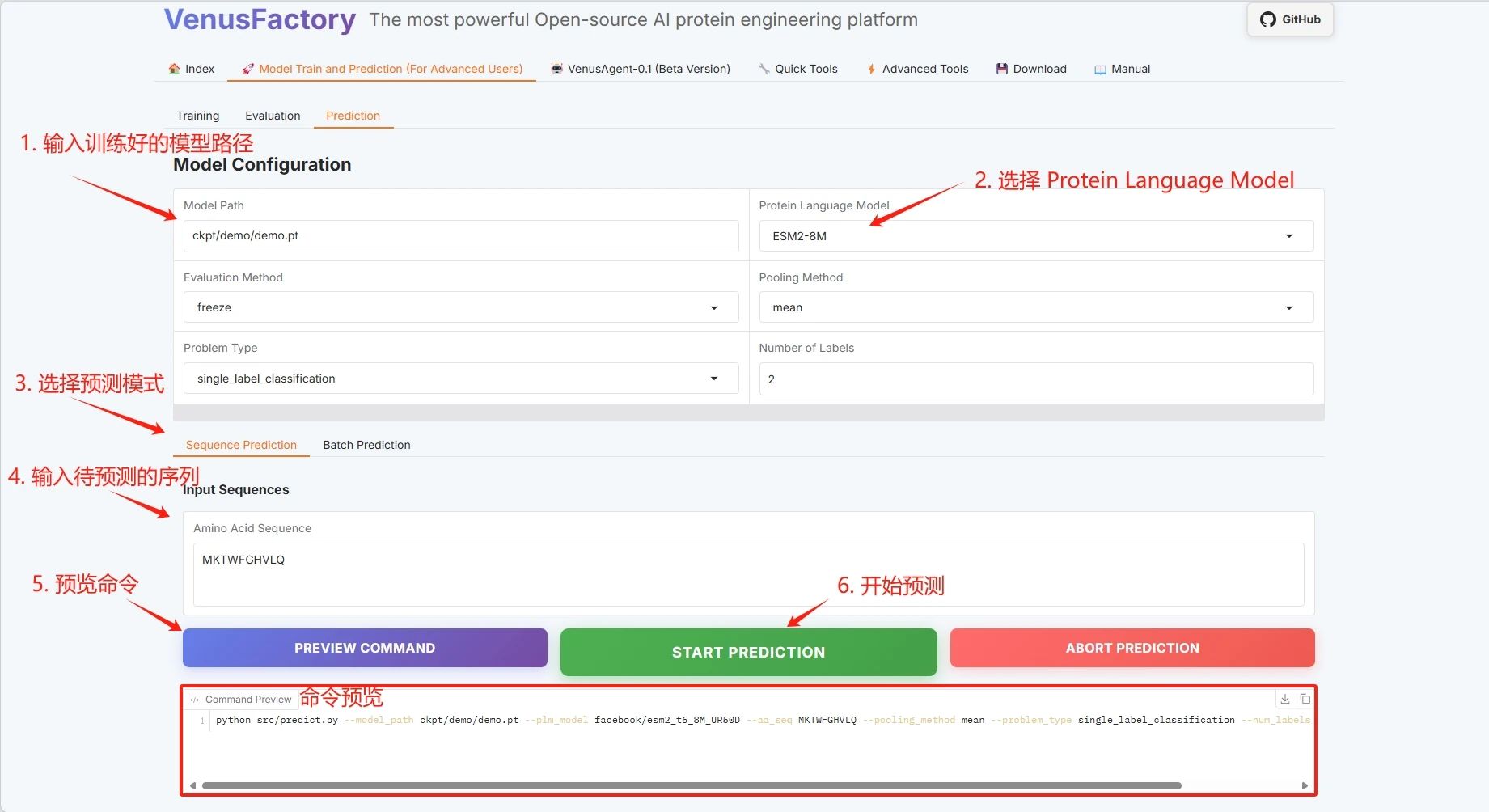
Protein sequence example: MKTWFGHVLQ
VenusAgent Showcase
Click the VenusAgent module.
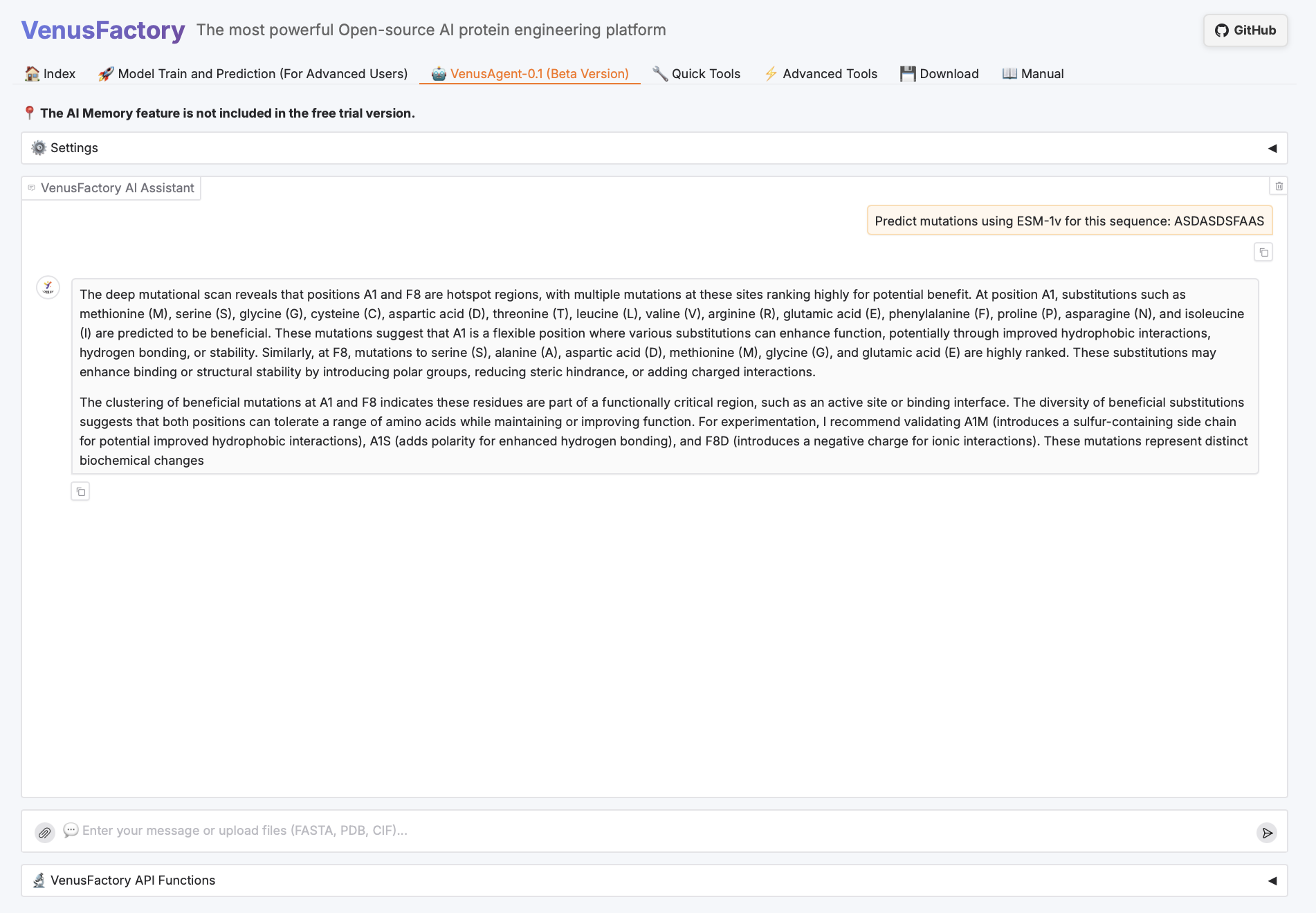
Since VenusAgent requires DeepSeek large models, this tutorial provides two calling methods: inputting the API key yourself or using the DeepSeek-R1-70B model deployed on the platform. You can choose different graphics card experiences based on the required functions. The card selection instructions are as follows:
* If using a single RTX 4090 graphics card, the VenusAgent function does not support the use of locally deployed large model services (using the DeepSeek API Key is unlimited).
* If you use dual RTX 4090 graphics cards, you cannot use other functions immediately (after 1-2 minutes) after using the VenusAgent function (there is no restriction when using the DeepSeek API Key).
* If using dual RTX A6000 graphics cards, VenusAgent functions are unlimited.
* Users can enter the DeepSeek API Key. If not, the default is to use the large model service deployed locally in the tutorial. When using the local large model service, the response time for the first conversation is approximately 2-3 minutes. Please be patient.
The above is a detailed tutorial on how to use the "VenusFactory Protein Engineering Design Platform". Everyone is welcome to come and experience it!
Tutorial Link:
Get high-quality papers and in-depth interpretation articles in the field of AI4S from 2023 to 2024 with one click⬇️