Command Palette
Search for a command to run...
Yu Xiang's Research Group at Shanghai Jiao Tong University Released a Transferable Deep Learning Model to Identify Multiple Types of RNA Modifications and Significantly Reduce Computational Costs

In 2021, due to the loud call of Gao Fu, an academician of the Chinese Academy of Sciences, mRNA vaccines became famous overnight and became people's hope during the outbreak of the new coronavirus. Today, that special past has become history, but the RNA modification behind mRNA vaccines is still moving forward at a rapid pace.
The so-called RNA modification is an important type of post-transcriptional regulation that can be widely involved in various RNA post-transcriptional processing and metabolic pathways.
RNA modification deserves attention because it plays a vital biological function in the growth and development of eukaryotic organisms.For example, recent studies have found that the destabilization effect of N⁶-methyladenosine (m⁶A) in mammalian embryonic stem cells is related to a variety of diseases, and 5-methylcytosine (m⁵C) is related to the tolerance of rice to high temperature.
However, RNA has many types of modifications. So far, more than 160 types of modifications have been found in natural RNA. Previously, the nanopore direct RNA sequencing (DRS) technology developed by Oxford Nanopore Technologies (ONT) combined with deep learning methods can realize the modification identification of single bases.However, this method has difficulty in detecting multiple modification types simultaneously in a single sample.
In response to the above questions, the research group of Yu Xiang, a tenured associate professor at the School of Life Sciences and Technology of Shanghai Jiao Tong University, and the team of Yang Jun/Wang Hongxia from Shanghai Chenshan Botanical Garden published a research paper titled "Transfer learning enables identification of multiple types of RNA modifications using nanopore direct RNA sequencing" in Nature Communications.A transferable deep learning model TandemMod was developed to enable identification of multiple types of RNA modifications in DRS.
Research highlights:
* Under the condition of ensuring the same performance, significantly reduce the computing costs such as the amount of training set data and model training time
* TandemMod provides important technical support for the identification of various types of RNA modification sites and epitranscriptome studies in animals, plants and microorganisms
* TandemMod can also be used to detect artificially modified RNA such as RNA vaccines

Paper address:
https://www.nature.com/articles/s41467-024-48437-4
The open source project "awesome-ai4s" brings together more than 100 AI4S paper interpretations and provides massive data sets and tools:
https://github.com/hyperai/awesome-ai4s
Dataset: Targeted training with multiple datasets
In order to train and evaluate the performance of the TandemMod model, the research team used multiple data sets for experiments.
first,The research team used the ELIGOS in vitro transcription dataset generated by the Nookaew laboratory.Five base-level features (mean, median, standard deviation, signal length, and base quality) were calculated for six modified bases (m¹A, m⁶A, m⁵C, hm⁵C, m⁷G, and Ψ) and compared with the unmodified bases.
Secondly, the research team chose to study the performance of TandemMod based on the two most common modifications in eukaryotic mRNA, m⁵C and m⁶A.The researchers trained the TandemMod m⁵C model on the Curlcake dataset.The dataset is derived from in vitro transcribed sequences containing all possible 5-mers and is divided into training and testing sets in a 4:1 ratio.
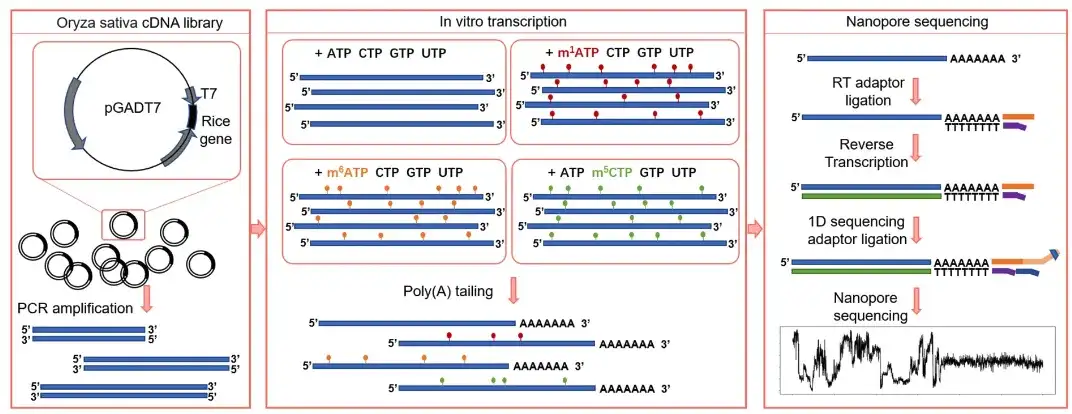
At the same time, in order to solve the problem that RNA transcribed from in vitro synthetic sequences cannot cover the entire range of natural sequences, the research team performed in vitro transcription on a rice cDNA library containing a T7 promoter, and obtained thousands of transcripts with different modification tags. After adding polyA tails, four training sets (m¹A, m⁶A, m⁵C, and unmodified bases) were constructed through DRS.It is called the In Vitro Apparent Transcriptome Dataset (IVET).
Model Architecture: A Deep Learning Framework
Based on this, the research team used the electrical signals assigned to every 5 bases and their statistical characteristics as input to train the transfer learning model TandemMod, which can simultaneously detect multiple types of RNA modifications.
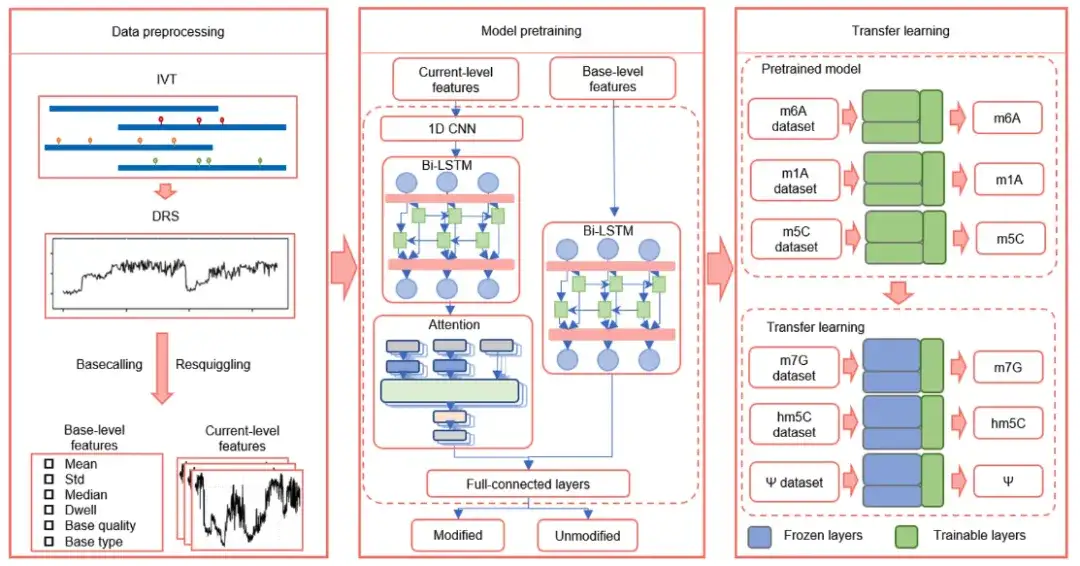
As shown in the figure above,TandemMod consists of data preprocessing, model pretraining, and transfer learning.
Among them, model pre-training consists of 4 main components:
* One-dimensional convolutional neural network (1D-CNN) to extract local features of the original current intensity signal;
* Bi-LSTM (Bi-LSTM), which is used to capture long-term correlations between adjacent signals and improve the ability to understand context in a longer process;
* Attention mechanism, which is used to weight the importance of each feature at different times and improve the model's ability to capture important signals;
* The classifier of the fully-connected layers is responsible for making predictions based on the combined information of all features.
In addition, to verify whether transfer learning can be applied to DRS data to detect multiple types of RNA modifications,The researchers trained TandemMod on the IVET m5C dataset and obtained a pretrained model.In the TandemMod model, the top layer acts as a feature extractor and the bottom layer acts as a classifier. The researchers froze the top layer of the pre-trained model and retrained the bottom layer on the ELIGOS training set (hm5C, m7G, Ψ, and I) to minimize the classification error.

After 2 epochs, all models achieved high accuracy.The ROC-AUCs of hm⁵C, m⁷G, Ψ, and I reached 0.98, 0.95, 0.96, and 0.97, respectively. As shown in Figures a, b, c, and d above.
Experimental results: TandemMod significantly reduces the amount of training set data and model training time
In the experimental stage, the research team compared the TandemMod model with classic machine learning algorithms to evaluate its performance, including XGBoost, support vector machine (SVM) and k-nearest neighbor (KNN). In the case of Curlcake test dataset m⁶A recognition,TandemMod outperforms other algorithms with an accuracy of 0.90.Similarly, for the identification of m⁵C, TandemMod achieved an accuracy of 0.95, and this comparison highlights the effectiveness of TandemMod in identifying modifications using DRS data.
TandemMod also showed better superiority than tombo and xPore in identifying samples with different modification rate levels in vivo.This indicates that TandemMod can accurately predict samples with different modification rates without the need for negative control samples.
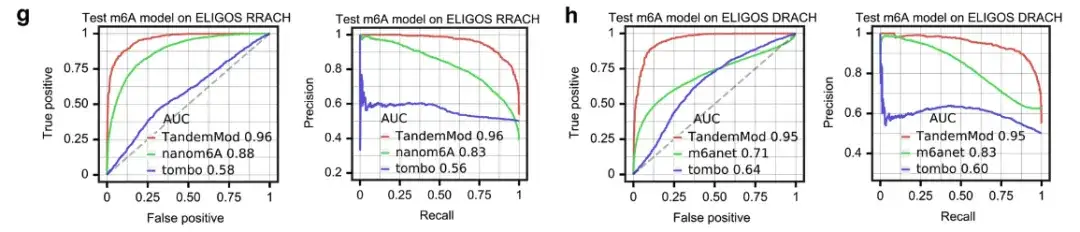
In addition, the research team also compared the TandemMod m⁶A model with tombo, nanom6A and m6Anet, as shown in the figure above.
On the ELIGOS rash (RA or G, HA, or C or U) motif, the ROC-AUCs of TandemMod, nanom6A, and tombo were 0.96, 0.88, and 0.52, respectively. On the ELIGOS DRACH (DA, G, or U) motif, the ROC-AUCs of TandemMod, m6Anet, and tombo were 0.95, 0.71, and 0.64, respectively.
These results indicate thatTrained using the in vitro DRS dataset, TandemMod provides the most accurate read-level predictions among existing tools.
The research team verified the classification performance, required training data, and computing resource utilization of the TandemMod m⁵C model in m⁶A detection, and compared it with the TandemMod m⁶A model of the standard instance. The results show that transfer learning can significantly reduce the cost of training set data volume and model training time while ensuring the same performance.
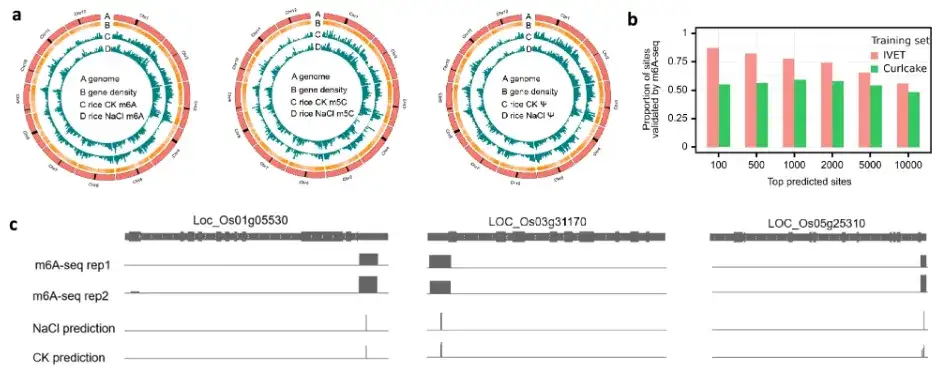
Finally, the research team tested the ability of the TandemMod model to be extended to new species for DRS data sequencing, and further verified the reliability of TandemMod using human cell lines (2 modification enzyme knockout samples and 5 wild-type samples). At the same time, the research team also used TandemMod to map the epigenetic modification maps of m⁶A, m⁵C, and Ψ in rice seedlings under high salt stress, and revealed the co-modification of m⁶A and m⁵C in mRNA and the changes in their modification rates under high salt conditions. As shown in the figure above.
RNA modification opens new doors to explore life
Throughout the ages, people have never stopped exploring life. After the RNA world hypothesis was proposed, the argument that RNA is the origin of life has undoubtedly become one of the most convincing answers. Since the first RNA modification was discovered in 1960, it has long been a top priority in the scientific research community and has maintained a high level of attention in recent years.
In addition to Yu Xiang's research group and Yang Jun/Wang Hongxia's team in this paper, as well as the ONT company mentioned in the article, there are many more teams and companies that are also conducting RNA modification research.
For example, in 2021, Professor Meng Jia's team from Xi'an Jiaotong-Liverpool University published an article titled "Attention-based multi-label neural networks for integrated prediction and interpretation of twelve widely occurring RNA modifications" in the journal Nature Communications.
Paper address:https://www.nature.com/articles/s41467-021-24313-3
The article mentions a model MultiRM based on a multi-label deep learning framework with attention mechanism.Not only can 12 widely existing transcriptome sites be predicted simultaneously, but key sequences in the prediction process are also extracted and analyzed, revealing a strong correlation between different types of RNA modifications, which helps to better comprehensively analyze and understand sequence-based RNA modification mechanisms.

Coincidentally, in a 2021 paper titled "Identification of differential RNA modifications from nanopore direct RNA sequencing with xPore" published in Nature Biotechnology,The research team used xPore to identify RNA modifications with high precision from Direct RNA-seq data and analyze differential modification and expression from a single high-throughput experiment.
Paper address:https://www.nature.com/articles/s41587-021-00949-w
These studies are helping us to further open the door to the RNA world, allowing us to further explore the "true meaning of life." Although there are still many bottlenecks to be overcome in the progress of various studies, the continuous challenges of "pioneers" have already made the door to RNA research more open.
References:
1. https://news.sjtu.edu.cn/jdzh/2