Command Palette
Search for a command to run...
Online Tutorial | David Baker's Team open-sources RFdiffusion3, Achieving a Generative Breakthrough in all-atom Protein design.

In recent years, significant progress has been made in the design of novel functional proteins using generative deep learning methods. Currently, most methods, including RFdiffusion (RFD1) and BindCraft, use protein representation at the amino acid residue level and have been able to successfully design protein monomers, assemblies, and protein-protein interaction systems. However, their resolution is still insufficient to accurately design structures that interact specifically with non-protein components (such as small molecule ligands and nucleic acids) through side-chain interactions.
While RFdiffusion2 (RFD2) has overcome this limitation to some extent, its diffusion process is still limited to the residue level and it is difficult to extend further to form additional side chain interactions with non-protein components.Existing research suggests that atomic-level diffusion processes can be used to generate protein backbones and can be extended to side-chain modeling, but these attempts have not yet achieved effective modeling of interactions between non-protein components.
Based on this,Nobel laureate David Baker's team has developed RFdiffusion3 (RFD3), which can generate three-dimensional conformations of proteins in structures composed of ligands, nucleic acids, and other non-protein atoms.Because this model explicitly models all polymer atoms, it can handle complex atomic-level constraints more easily and efficiently in tasks such as enzyme design. RFD3's native all-atom architecture also greatly simplifies the specification of atomic-level constraints, providing precise control over hydrogen bonds, ligand contacts, and nucleic acid interactions.
Unlike AlphaFold3 (AF3), which relies on a computationally intensive Pairformer module to extract information such as distance from the input sequence, the research team designed a more lightweight information extraction module.This allows RFD3 to drastically reduce the number of layers in the Pairformer from 48 to only 2, thereby significantly reducing computational overhead. The final model contains only 168 million trainable parameters.The research team demonstrated the broad applicability of RFD3 by designing and experimentally characterizing DNA-binding proteins and cysteine hydrolases. RFD3 can rapidly generate protein structures guided by complex atomic-level constraints based on any non-protein atomic environment, which will further expand the functional range that protein design can achieve.
"RFdiffusion3: Protein Design Model" is now available on the HyperAI website (hyper.ai) in the "Tutorials" section. Download and experience it with one click!
To celebrate the New Year, HyperAI is offering computing power benefits to everyone.New users can get 2 hours of NVIDIA credit by using the redemption code "2026 Happy New Year" after registration. GeForce RTX 5090 Usage duration,Limited quantities available, grab your holiday perks now!
Tutorial Link:
Demo Run
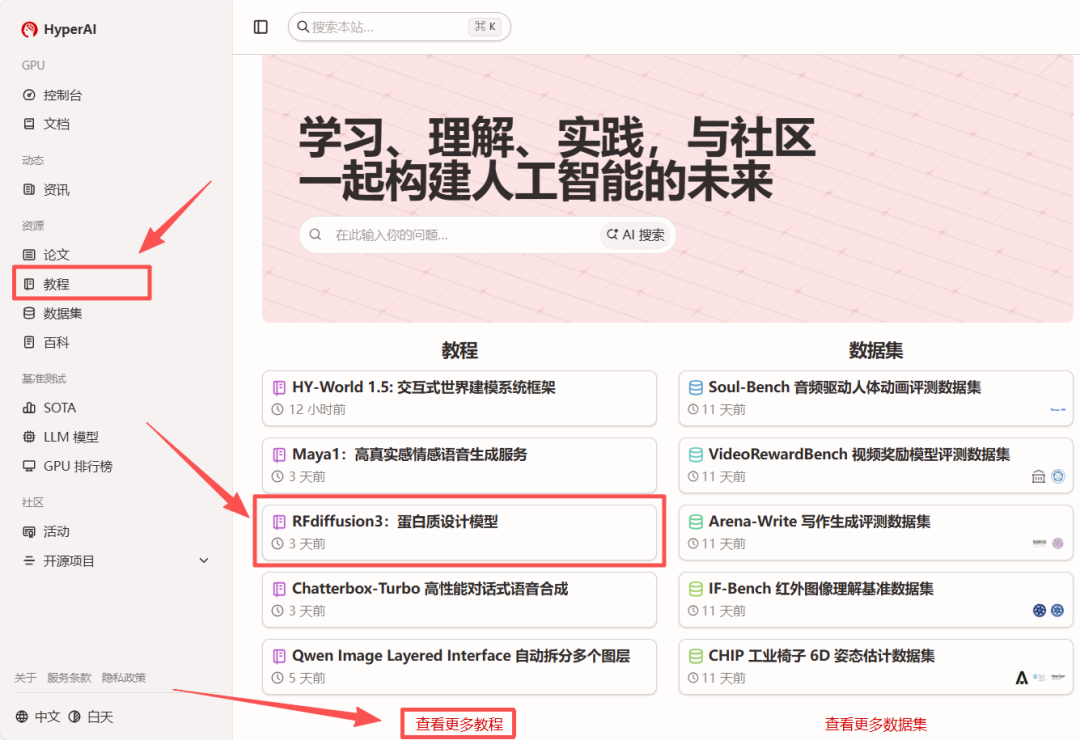
1. After entering the hyper.ai homepage, select "RFdiffusion3: Protein Design Model", or go to the "Tutorials" page to select it. Then click "Run this tutorial online".
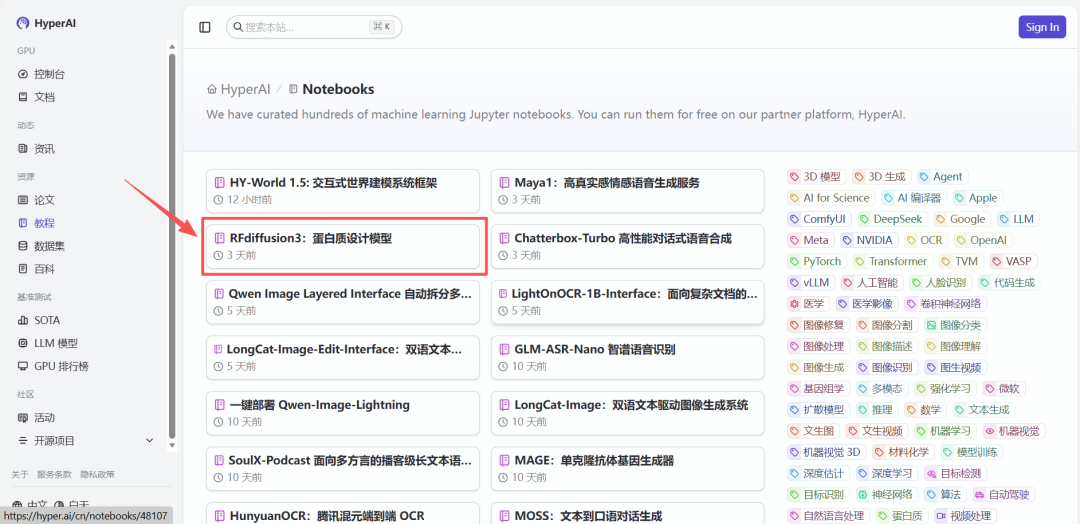
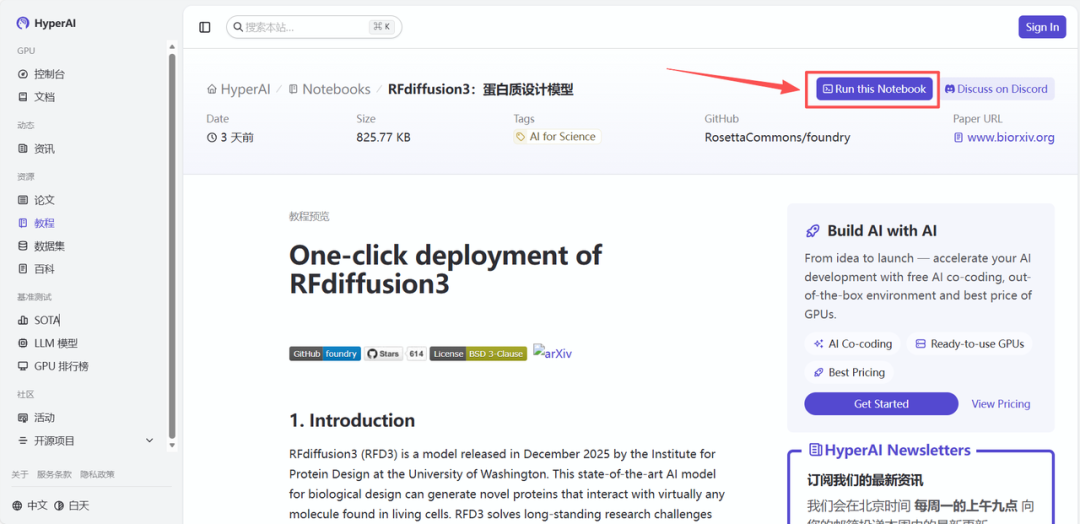
2. After the page redirects, click "Clone" in the upper right corner to clone the tutorial into your own container.
Note: You can switch languages in the upper right corner of the page. Currently, Chinese and English are available. This tutorial will show the steps in English.
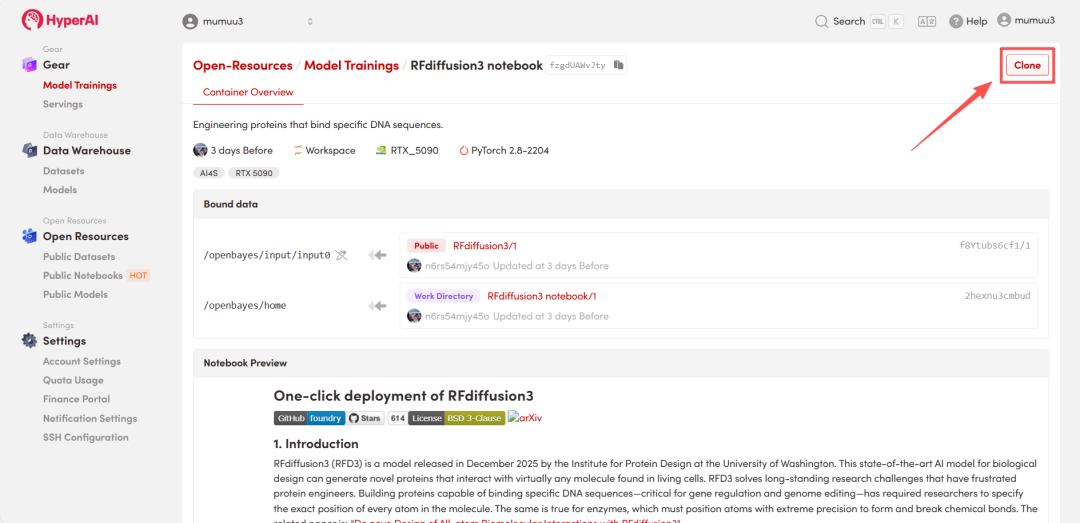
3. Select the "NVIDIA GeForce RTX 5090" and "PyTorch" images, and choose "Pay As You Go" or "Daily Plan/Weekly Plan/Monthly Plan" as needed, then click "Continue job execution".
HyperAI is offering a registration bonus for new users: for just $1, you can get 20 hours of RTX 5090 computing power (originally priced at $7), and the resources are valid indefinitely.
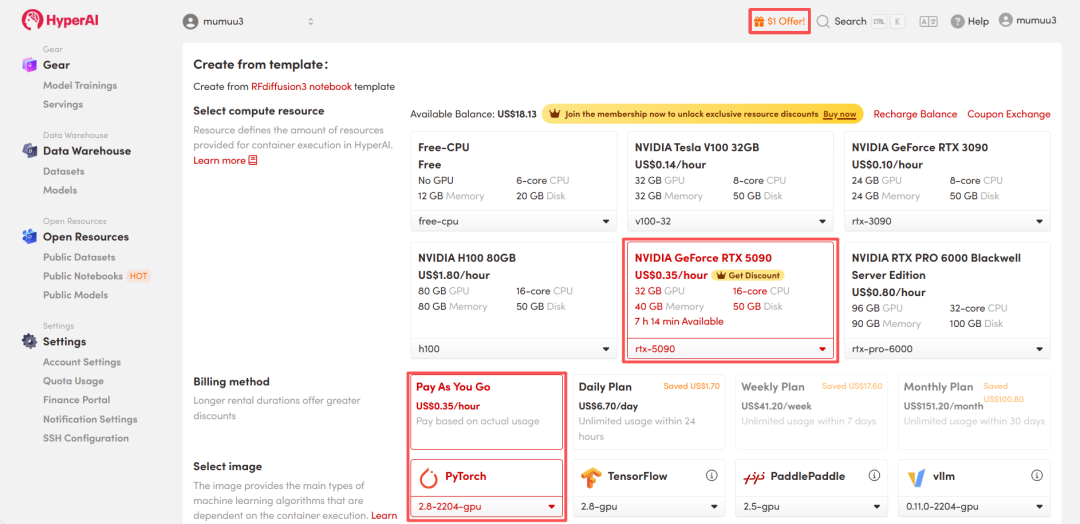
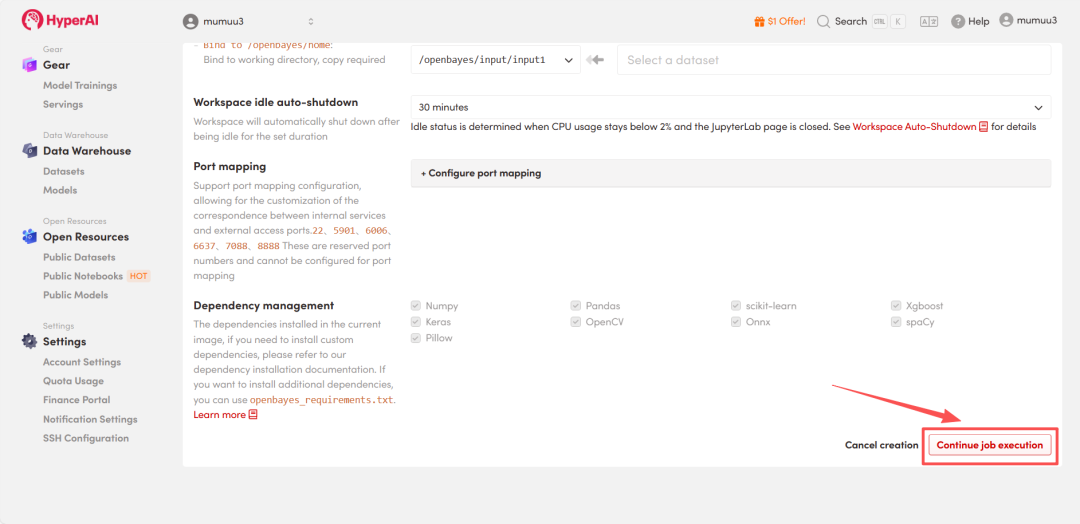
4. Wait for resources to be allocated. Once the status changes to "Running", click "Open Workspace" to enter the Jupyter Workspace.
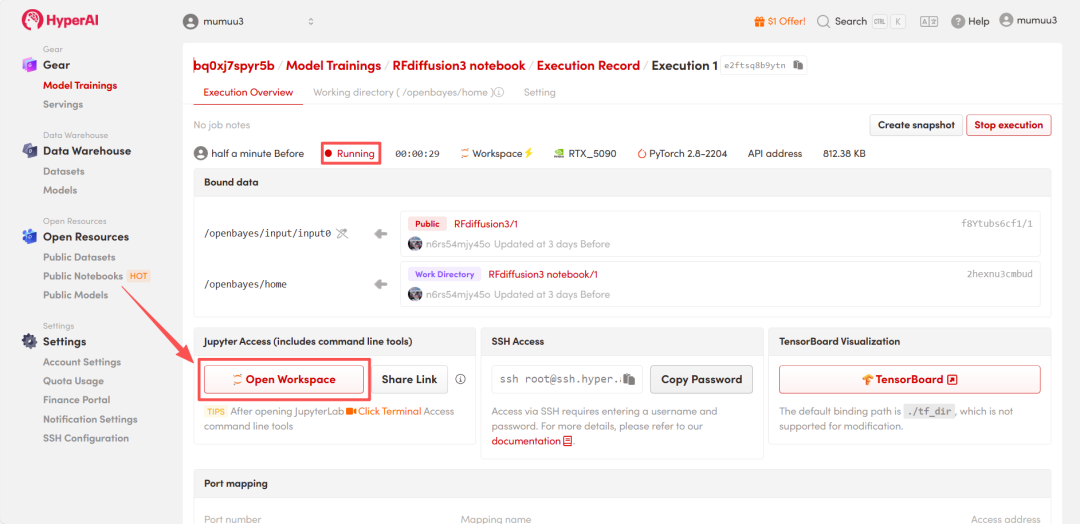
Effect Demonstration
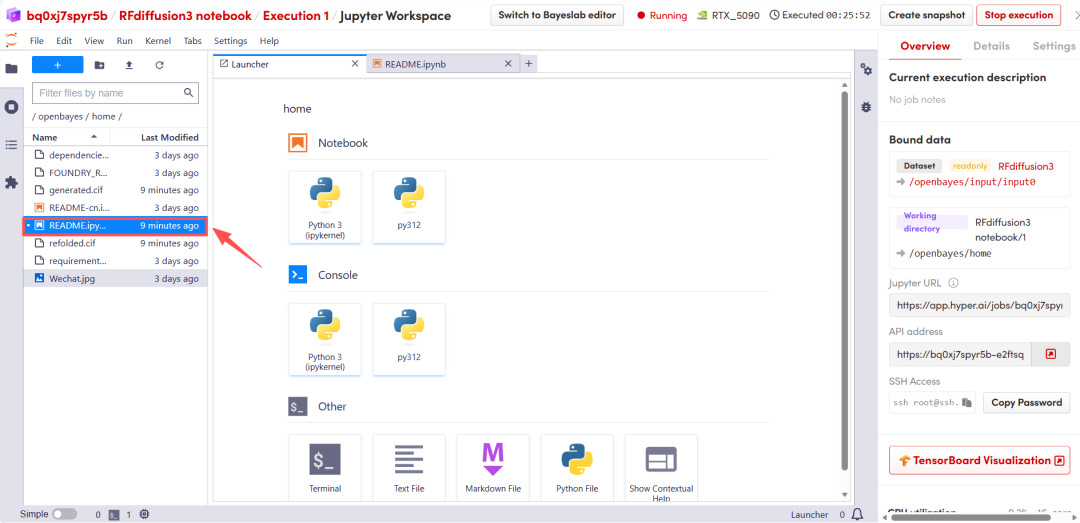
After the page redirects, click on the README page on the left, and then click Run at the top.
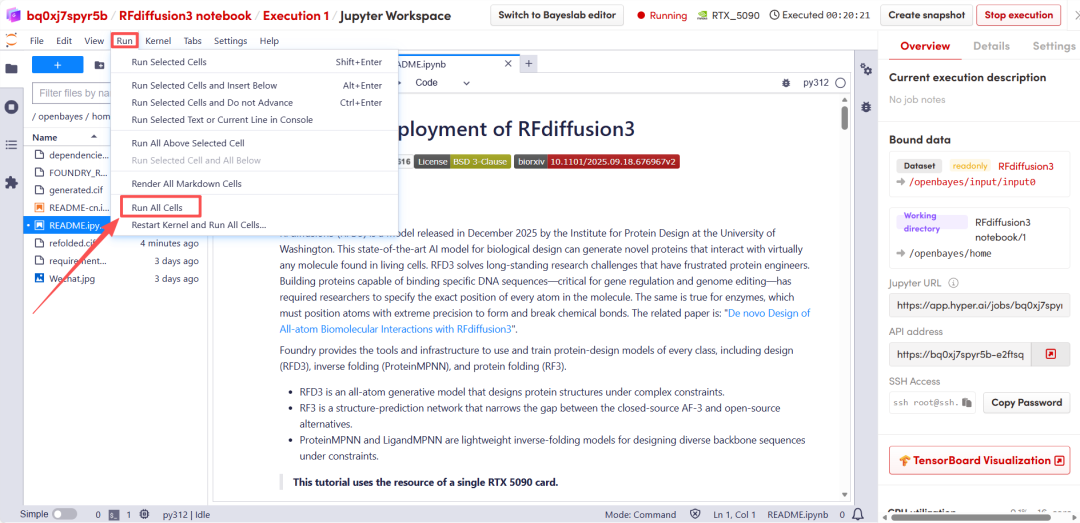
After a short wait, scroll down to display the structural prediction results from RFD3.
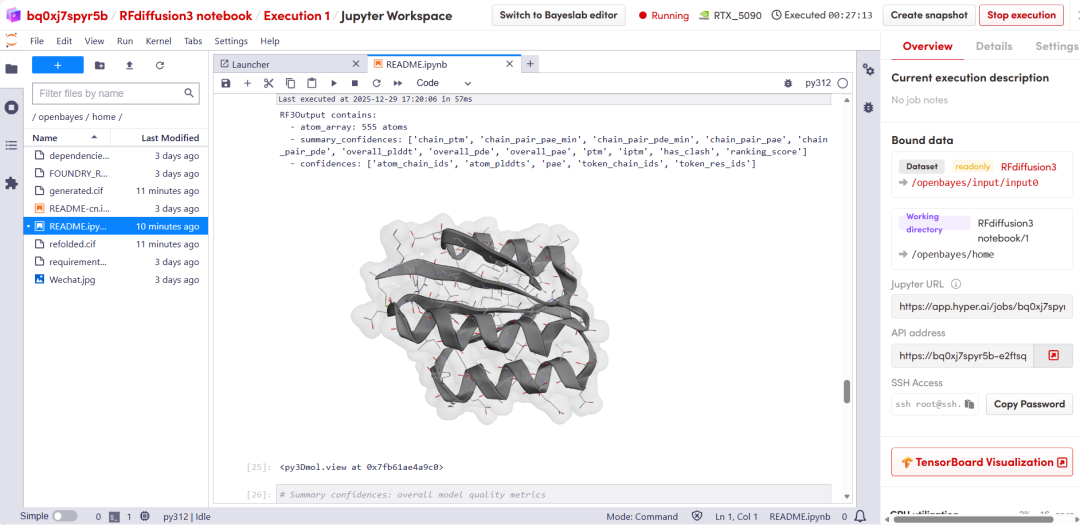
The above is the tutorial recommended by HyperAI this time. Everyone is welcome to come and experience it!
Tutorial Link: