Command Palette
Search for a command to run...
A Carnegie Institution Interdisciplinary Team Successfully Captured Evidence of Life Dating Back 3.3 Billion Years Using a Random Forest Model Based on 406 samples.

Decoding the organic molecules buried deep beneath the Earth's surface in ancient rock layers is crucial for understanding Earth's history and studying the evolution of life. These potential witnesses to life activities can not only unravel the mysteries of the birth of life on Earth, especially clarifying the origin of photosynthesis and the connection between atmospheric oxidation, but also fill gaps in the timeline of life's evolution, providing key clues for understanding the formation of early Earth's ecosystems. However, unlike large organisms that form visible fossils, these "witnesses" have long since disappeared without a trace due to geological erosion.Therefore, identifying traces of life from highly degraded organic remains has become a major challenge in the fields of paleontology and earth science.
For a long time, scientists have relied mainly on paleontological fossil morphology and isotope analysis to explore early life. However, these methods are often limited by the preservation state of the samples: for example, clear records of complex molecules such as lipids and porphyrins can only be traced back to about 1.6 billion years ago, far shorter than the time of origin of life revealed by other evidence. Furthermore, the origin of organic molecules in Archean rocks is unclear, and the boundary between biogenic and abiotic origins is difficult to determine, all of which have kept many key discoveries at the speculative stage.
In order to break this deadlockLed by the Earth and Planetary Laboratory of the Carnegie Institution for Science, and in collaboration with a cross-disciplinary team comprised of numerous universities and research institutions worldwide, a "technology convergence" solution has been proposed.They first used pyrolysis–gas chromatography–mass spectrometry (py-GC-MS) for analysis, and then used supervised machine learning methods to classify and discriminate the analyzed data, thereby capturing ancient traces of life in the chaotic molecular fragments.
Experiments show that the model integrating these technologies exhibits better-than-expected results. It can accurately distinguish between modern organic matter and meteorite/fossil organic matter with a resolution of 1001 TP3T, and can distinguish between fossil plant tissue and meteorite organic matter with an accuracy of 971 TP3T. More importantly, when the team applied it to unknown samples, the model was able to successfully identify evidence of biogenic molecular assemblages in Paleoarchean and Neoarchean rocks from 3.33 billion years ago and 2.52 billion years ago. This provides new methodological support for exploring earlier and less surviving traces of life.
The related research, titled "Organic geochemical evidence for life in Archean rocks identified by pyrolysis–GC–MS and supervised machine learning," was published in the Proceedings of the National Academy of Sciences (PNAS).
Research highlights:
* The proposed technology fusion approach breaks through traditional limitations, overcoming the core challenge of distinguishing molecules after degradation by combining pyrolysis gas chromatography-mass spectrometry with machine learning.
* The research sample covers a wide range, from modern life to rocks from billions of years ago, from terrestrial organisms to extraterrestrial meteorites, providing a comprehensive comparison for model training.
* Experiments show that this method is both scientifically sound and forward-looking, not only verifying the existence of life traces in Archean rocks, but also providing a new method for exploring other unknown life traces.

Paper address:
https://www.pnas.org/doi/10.1073/pnas.2514534122
Follow our official WeChat account and reply "pyrolysis gas chromatography" in the background to get the complete PDF.
Dataset: 406 samples, covering a wide range, providing a comprehensive comparison for the model.
The research team analyzed a total of 406 natural and synthetic samples containing a range of organic molecules, covering ancient and modern, biological and abiotic sources, spanning from approximately 3.8 billion years ago (Archean) to 10 million years ago (Neogene). The sample types included sedimentary rocks (141 pieces), fossils (65 samples), modern organisms (123), meteorites (42, of which 39 were carbonaceous chondrites), and laboratory-synthesized organic molecules (35 groups), providing a rich and diverse data foundation for machine learning analysis.
Of these 406 samples, 272 were clearly divided into 9 categories based on phylogenetic relationships and physiological characteristics, and were used for supervised machine learning training (75%) and testing (25%), as shown in the figure below:
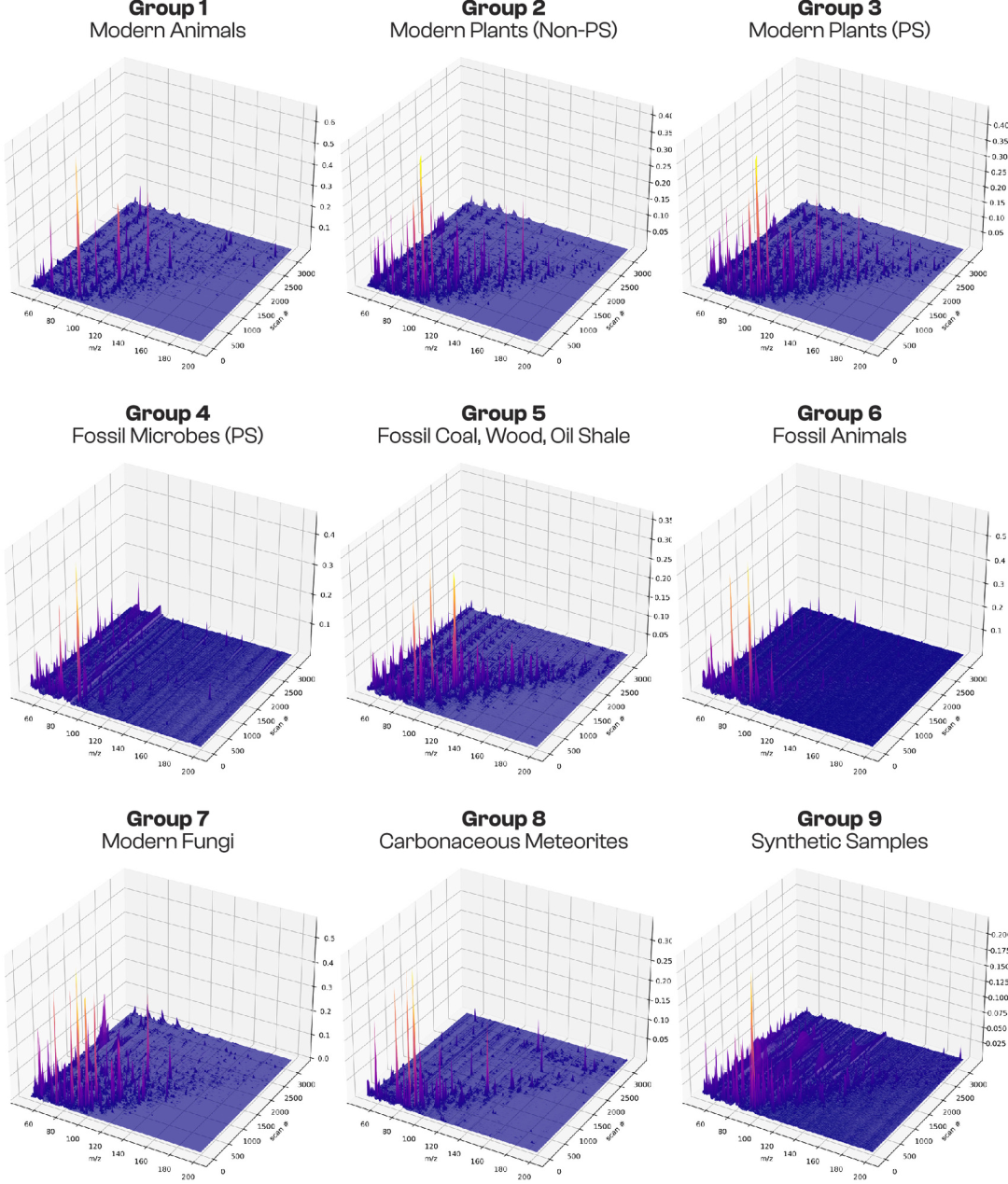
* Modern animals:Organic molecular characteristics of modern non-photosynthetic heterotrophic organisms were derived from a variety of recently deceased invertebrates and vertebrates. The sample size was 21.
* Modern plants (non-photosynthetic tissues):This study included non-photosynthetic tissues and secretions from plant roots, seeds, flowers, fruits, and sap, representing the molecular differences among different functional tissues of plants. The sample size was 40.
* Modern plants (photosynthetic tissue):The study primarily focused on leaves and other photosynthetic tissues, serving as a modern reference for the characteristics of photosynthetic biomolecules. The sample size was 36.
* Sedimentary rocks containing photosynthetic cyanobacteria/algal fossils:Organic residues enriched by acid dissolution with hydrochloric acid (HCl) and hydrofluoric acid (HF) in shale or flint, and the rocks possess reliable morphological evidence of cyanobacteria or algal fossils, serving as a molecular record of ancient photosynthetic microorganisms. The sample size was 24.
* Petrified wood, coal, and oil shale:The samples are mainly from the Phanerozoic Eon (<541 million years ago), but also include complex hydrocarbon-rich sediments from the Proterozoic, such as shungite and anthraxolite, representing the molecular preservation characteristics of ancient higher plants and hydrocarbons. The total number of samples is 49.
* Animal fossils:All samples are from the Phanerozoic Eon, including carbonized remains of fish fossils and trilobite fossils, as well as shell-binding proteins extracted from the shells of Miocene gastropods, representing organic molecular remains of ancient animals. There are nine samples in total.
* Modern fungi:It includes a variety of wood-decaying fungi and yeasts, filling gaps in the molecular data of non-plant and non-animal groups in eukaryotes. The sample size is 16.
* Meteorite:The samples were primarily carbonaceous chondrites (39 in total), which underwent chemical dissolution and enrichment of organic molecular assemblages, serving as a clear reference for non-biological organic sources. A total of 42 samples were collected.
* Laboratory-synthesized samples:The study used assemblages of organic molecules obtained through laboratory synthesis processes such as the Maillard reaction and the Formose reaction to simulate the molecular characteristics of abiotically derived organic substances. The sample size was 35.
besides,The research team also set up two additional auxiliary class samples for specific machine learning models.To distinguish between photosynthetic and non-photosynthetic organisms, a total of three samples were used. Two modern cyanobacteria samples were used to supplement the data on photosynthetic prokaryotes. One modern halophilic bacterium (Halobacter) sample was used to supplement the data on non-photosynthetic archaea.
Finally, the remaining 131 samples were mainly acid-soluble enriched residues from Archean or Proterozoic sedimentary rocks rich in organic matter. The origin and physiological characteristics of the organic molecules in these samples are unknown or controversial, but this also provides a new classification test field for verifying the application of machine learning analysis in this experiment.
Research Methods and Models: Deep Integration of py-GC-MS and Machine Learning
This experiment can be summarized into four main steps:
* The first step involved collecting 406 different carbon-containing samples from various modern and ancient, biological and abiotic sources;
* The second step is to extract carbonaceous macromolecules from meteorites and ancient sedimentary rocks;
* The third step involves analyzing each sample using pyrolysis gas chromatography coupled to electron impact ionization mass spectrometry;
* Step 4: Use the data from the experimental sample analysis subset (machine learning method) to train a supervised random forest model.
The most important aspect of this method is the "technical integration" of py-GC-MS analysis techniques with machine learning methods.
First, there is the analytical technique.In this experiment, the research team used a CDS 6150 thermal probe coupled with an Agilent 8860 series gas chromatograph and an Agilent 5999 quadrupole mass spectrometer. Chromatographic separation was performed using an Agilent 30 M 5% phenyl PDMS column. The pyrolysis products were immediately swept onto the gas chromatographic column by helium for analysis. The specific procedures are as follows:
* Pyrolysis:Researchers loaded samples (10-100 μg) into preheated (burned in air at 550°C for 3 h) quartz tubes, then inserted them into a thermal probe coil for flash pyrolysis, heating them to 610°C at a rate of 500°C/s and holding them for 10 s.
* Chromatography:The initial temperature was 50℃, held for 1 minute, then increased to 300℃ at a rate of 5℃/min, held for 15 minutes. Ultra-high purity helium (UHP 5.5 grade) was used as the carrier gas.
* Mass spectrometry:It operates in electron ionization (EI) mode with an ionization energy of 70 eV at 250℃, with a scan range of m/z 45-700, a scan rate of 0.80 s/decade, and an inter-scan delay of 0.20 s.
To avoid interference from small molecule volatiles (such as CO₂ and H₂O), MS data were not collected for the first two minutes of the experiment. Furthermore, the experiment required excluding signals from elution regions of common contaminants in the chromatogram (such as palmitic acid and stearic acid). Each sample was converted into a two-dimensional matrix (3,240 elution time intervals x 150 m/z values), and the signal intensity of 489,240 elements was recorded as a function of mass and retention time. After standardization and smoothing, 8,149 effective features were ultimately retained.
Secondly, model selection was used. This experiment employed the random forest method.This is an ensemble classification method with high accuracy, low computational cost, and interpretability. It reduces the risk of overfitting by constructing multiple decorrelation decision trees. The model adopts the random forest model mentioned by Leo Breiman in "Random Forests".
Researchers used two validation strategies on the trained machine learning model. First, stratified random sampling was used with a training set of 75% and a test set of 25% to ensure that the proportion of each class of samples was consistent in the two groups. Then, the generalization ability of the model was evaluated by 10 repeated 10-fold cross-validation, and the average accuracy was calculated to reduce random error.
The experiment tested four models to distinguish between modern biogenic sources (plants and animals) and abiotic sources (meteorites + synthetic samples), ancient biogenic sources (sedimentary rocks of known biogenic origin) and abiotic sources, ancient biogenic sources (excluding petrified wood and coal) and abiotic sources, and photosynthetic and non-photosynthetic samples.
Experimental Results: Multi-model, multi-dimensional approach verifies the feasibility of technology integration.
In preliminary testing, researchers used a random forest model to classify 36 pairwise combinations of samples with 9 known attributes. Under conditions of relatively balanced sample sizes,Of the 36 tests, 25 had an accuracy of ≥ 90% on both the training and test sets, with 19 of them having an accuracy of ≥ 95%.All results are shown in the table below:
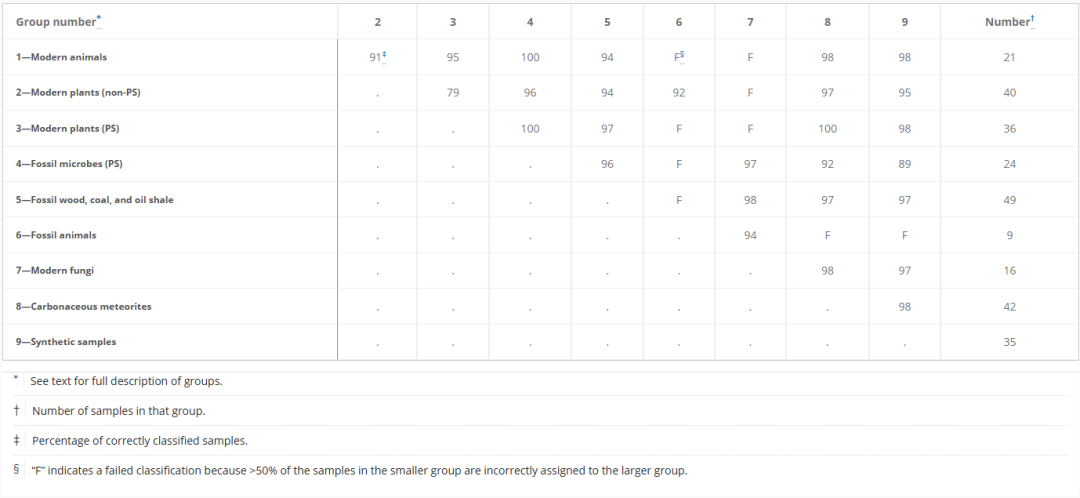
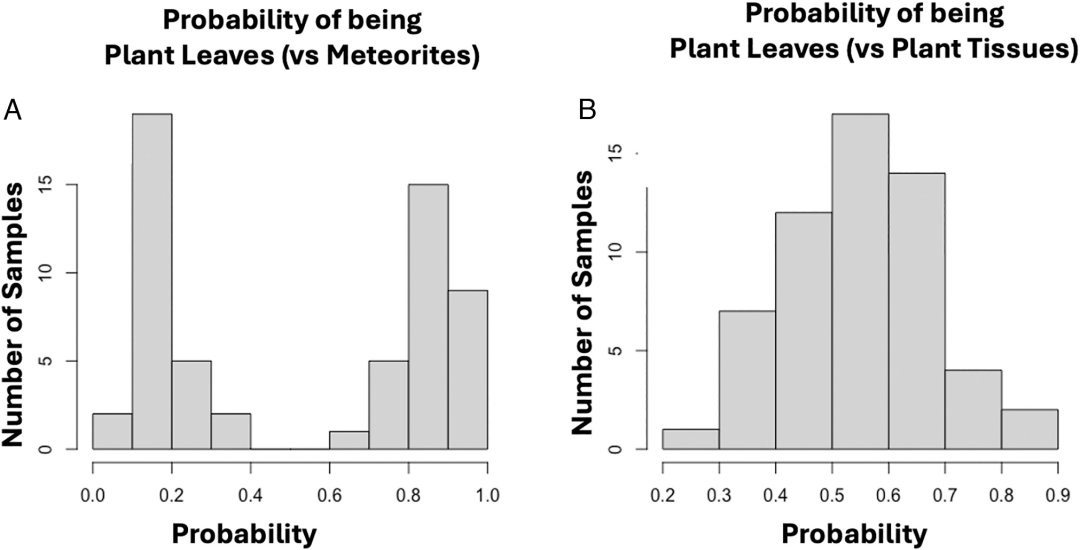
To further illustrate the method, the paper presents several case studies demonstrating the differences in efficiency and inefficiency across various cases. For example, in groups 3 and 8, namely modern plants (photosynthetic tissue) and meteorites,This method distinguished plants from meteorites with an accuracy of 100%.All samples had class probabilities > 0.6 or < 0.4, indicating significant differences in molecular characteristics. See Figure A below:
In addition, identifying biogenic and abiotic samples is a key objective of paleontology and astrobiology research. To this end, the research team constructed and compared three different random forest models to verify their ability to distinguish between biogenic and abiotic sources for different sample combinations.
Specifically, in model # 1, the research team tested the ability to distinguish between modern plants and animals and abiotic sources (meteorites and synthetic samples) in groups 1, 2, 3 and groups 8, 9, with sample numbers of 97 and 77, respectively.The overall accuracy rate reached 981 TP3T.The AUC value is 0.977 on the training set and 1.000 on the test set; the accuracy of 10-fold cross-validation is 98.3%.
Model # 2 was primarily used to validate the ability to distinguish between ancient biological samples and abiotic samples rich in organic matter. The control samples were from groups 4 and 5 and groups 8 and 9, containing 87 and 77 samples, respectively.Of the 87 biogenic ancient organic samples, 83 were correctly classified, achieving an accuracy rate of 95.1 TP3T.In addition, 70 of these samples (80%) had high confidence in biological origin classification probabilities, >0.6. 69 of the non-biological samples were correctly classified, achieving an accuracy of 90%; the AUC value was 0.924 on the training set and 0.926 on the test set; the 10-fold cross-validation accuracy was 92.7%.
When Model # 2 was applied to 109 ancient sedimentary rocks of unknown biogenic origin, 68 samples (61%) were found to have a biogenic origin classification probability > 0.50, and 32 samples were found to have a biogenic origin classification probability > 0.60.
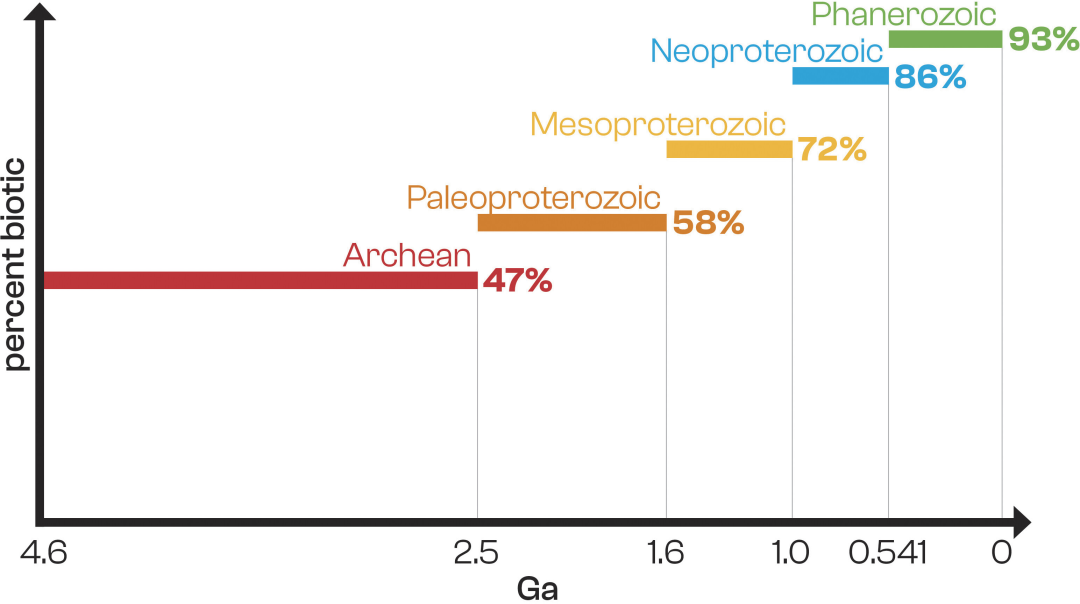
Furthermore, the results revealed a decreasing trend in the proportion of biogenic samples with geological age. Of the 82 Phanerozoic samples, 76 (93%) were biogenic, 43 (73%) were from the Proterozoic, and only 21 (47%) were from the Archean (45 samples). This shows a significant decrease in the percentage of biogenic samples with increasing age, possibly reflecting biomolecular degradation or abiotic organic input in the samples. (See figure below.)
Model # 3 is primarily used to verify the ability to distinguish between ancient biogenic and abiotic sources. The biogenic samples come from 89 shale and flint samples, including the fourth group of samples, while the abiotic samples remain the 77 samples from the eighth and ninth groups.All biological samples were correctly classified. 80% samples have a high confidence probability of biological cause classification (>0.60), and the accuracy of non-biological cause samples is 77%; the AUC value is 0.873 for the training set and 0.863 for the test set; the accuracy of 10-fold cross-validation is 91.6%.
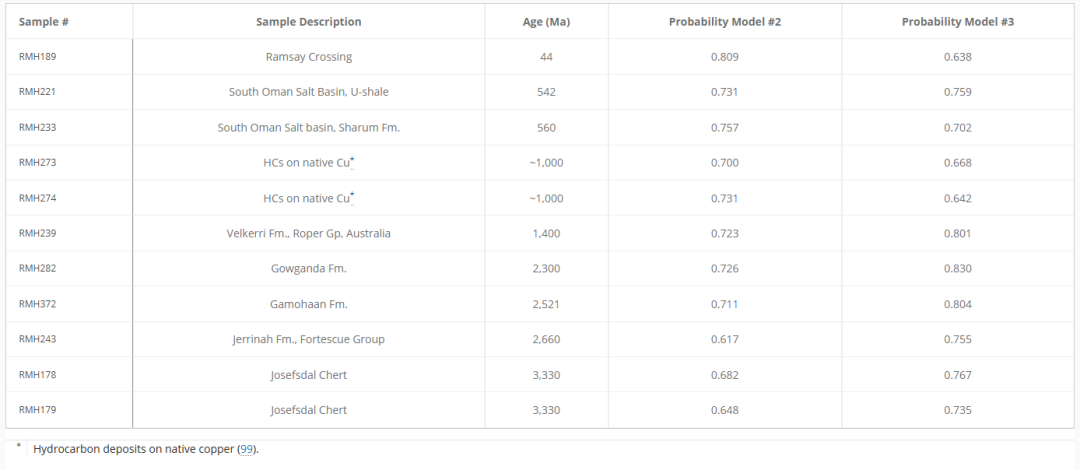
Furthermore, by combining Model # 2 and Model # 3,Researchers have identified 11 ancient samples as biologically derived, the oldest being the Josefsdal flint from the Barberton Greenstone Belt in South Africa, dating back 3.33 billion years.As shown in the following table:
Technological integration has become an important means of exploring the origins of life.
In recent years, global research teams have conducted numerous innovative explorations to address core challenges such as the identification of early life traces and the tracing of extraterrestrial organic matter. These studies also focus on the analysis of complex molecular mixtures, using algorithmic models to delve into biological characteristics that are difficult to capture using traditional analytical methods, laying a solid foundation for the feasibility of technological integration pathways and tracing the origins of life on Earth.
For example, the results of the Carnegie Institution for Science Earth and Planetary Laboratory, in conjunction with other different institutions, also used the methods mentioned above. These methods can be used to determine the biological origin of organic matter in planetary samples, as well as to identify traces of early life on Earth.This method combines pyrolysis gas chromatography-mass spectrometry measurements of terrestrial and extraterrestrial carbonaceous materials with machine learning classification methods.It achieved an accuracy of 90% in distinguishing between samples of non-biological origin and biological samples (including highly degraded biological samples), and accurately reflects the necessity of Darwin's biomolecular selection function.
Paper title: A robust, agnostic molecular biosignature based on machine learning
Paper address:https://www.pnas.org/doi/10.1073/pnas.2307149120
The integration of py-GC-MS and machine learning not only breaks the limitations of traditional methods in exploring early life, but also establishes a new paradigm at the intersection of paleontology and artificial intelligence. However, as seen in the aforementioned experiments and other studies, this technology-integrated approach still has room for optimization, providing direction for further in-depth research. It is believed that with continuous technological advancements, the future may enable humanity to gain a more intuitive and profound understanding of the origins of life, and even search for traces of extraterrestrial life.