Command Palette
Search for a command to run...
Online Tutorial | Computing Power Costs Plummet! Apple Launches Ml-simplefold, a Protein Folding Model Based on Stream matching.

In September 2025, Apple launched the lightweight protein folding prediction AI model Ml-simplefold. As the first protein folding model based on flow matching, it has been tested in authoritative benchmarks such as CAMEO22 and CASP14.After reducing the computational cost, SimpleFold still shows performance comparable to top models such as AlphaFold2 and RoseTTAFold2.At the same time, its smaller version, SimpleFold-100M, also performs competitively.
While traditional protein folding models offer impressive accuracy, they rely on a large number of domain-specific architectural designs and handcrafted features. The commonly used triangular update modules, explicit pair representation mechanisms, and multiple training objectives in these models make training computationally expensive. Furthermore, the architecture and hardware lack scalability, making it difficult to support the generation of diverse structures or ensemble prediction.
Aiming at the common defects of traditional methods,SimpleFold proposes a general generative framework based purely on Transformer, breaking the dependence of protein folding models on complex architectures:
* Based on Flow Matching technology, it skips complex modules such as multiple sequence alignment (MSA) and directly generates the three-dimensional structure of proteins from random noise, significantly reducing computational costs;
* Adopt a general architecture that abandons specific modules such as triangle update and pair representation, using only the standard Transformer and enhancing structure perception through adaptive layers;
* Structural constraints are added to give the model greater flexibility and physical consistency when generating protein three-dimensional structures. The 3B parameter model trained on 9M structural data can run smoothly on consumer-grade hardware, significantly reducing the computational threshold.
"Ml-simplefold: A Lightweight Protein Folding Prediction AI Model" is now available on the HyperAI Hyperneuron website (hyper.ai) in the "Tutorials" section. Run it with one click to experience the latest protein generation tool.
Tutorial Link:
Demo Run
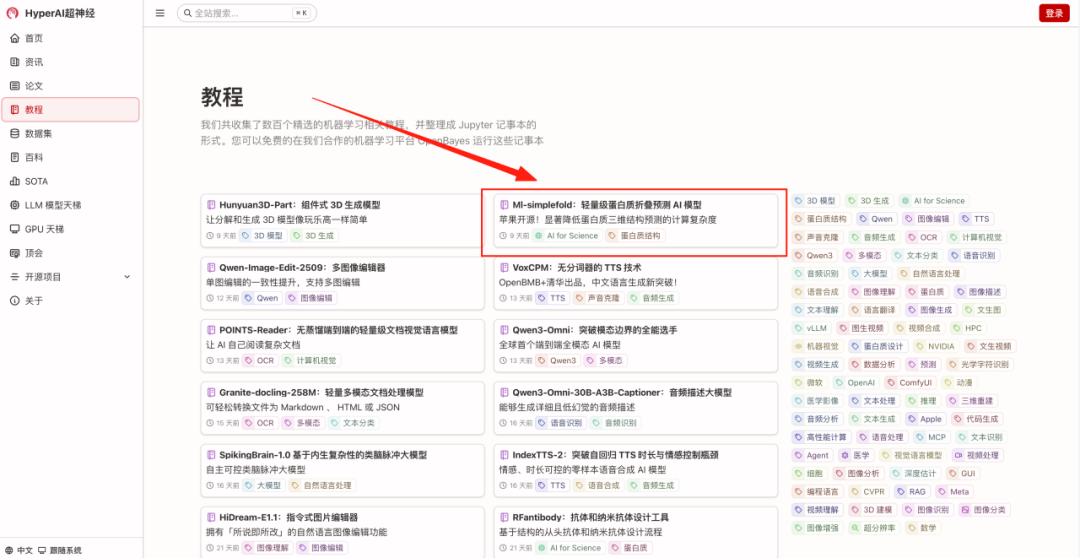
- Enter the URL hyper.ai in your browser. After entering the homepage, click the "Tutorial" page, select "Protein Folding Model Based on Flow Matching", and click "Run this tutorial online".
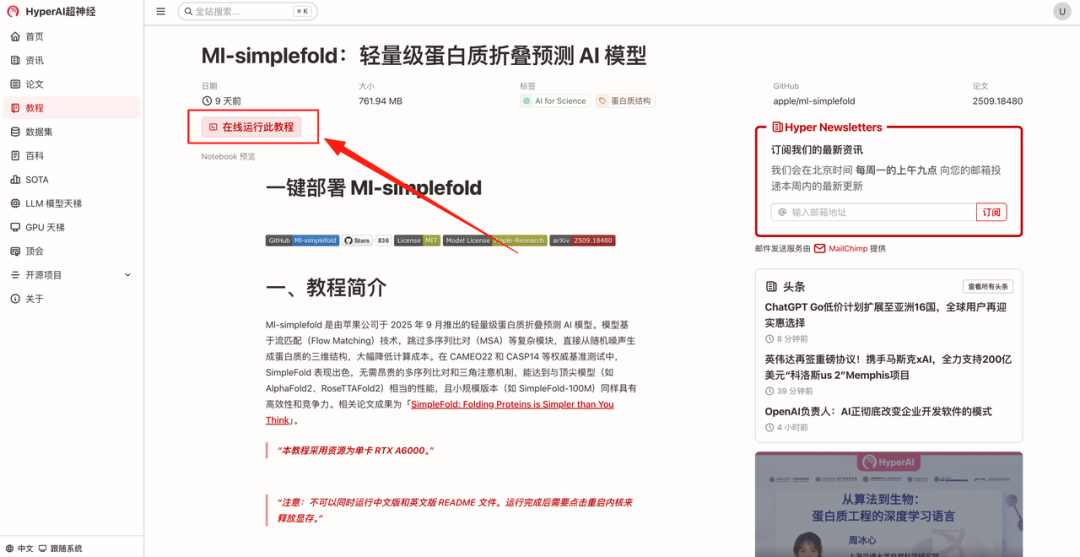
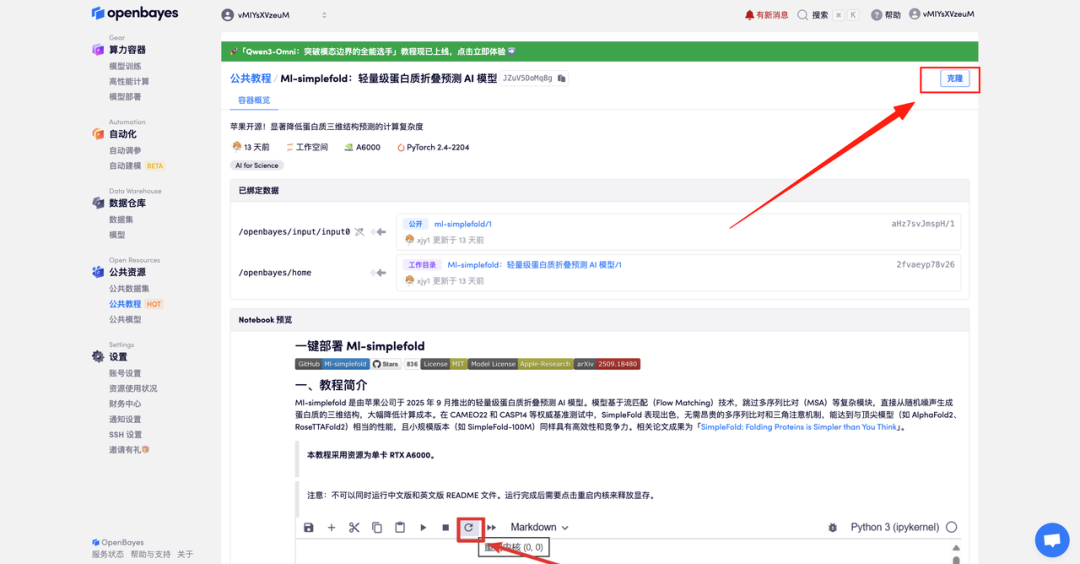
- After the page jumps, click "Clone" in the upper right corner to clone the tutorial into your own container.
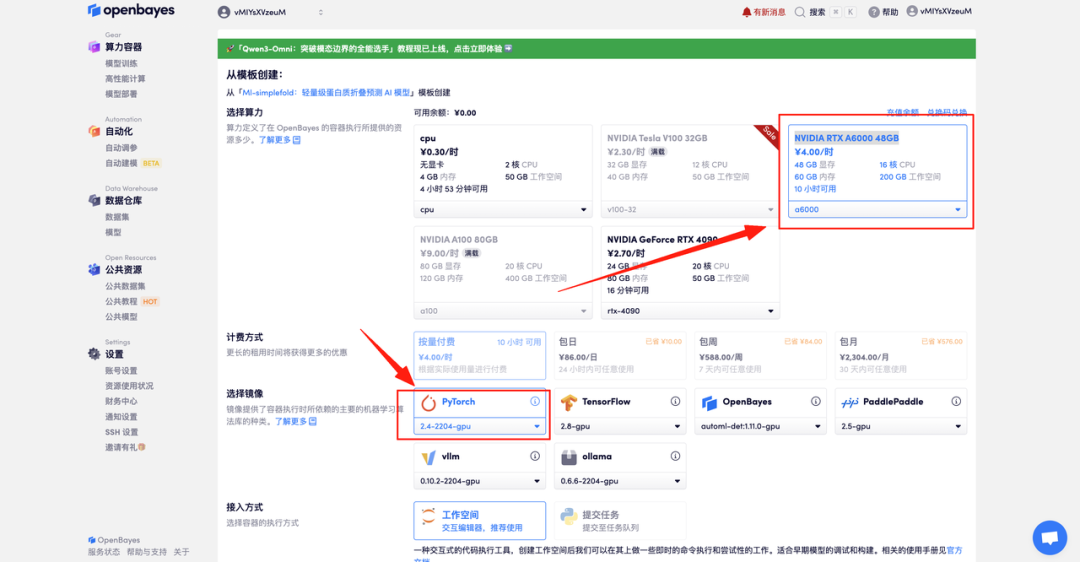
- Select the "NVIDIA RTX A6000 48GB" and "PyTorch" images and click "Continue." The OpenBayes platform offers four billing options: pay-as-you-go or daily/weekly/monthly plans. New users can register using the invitation link below to receive 4 hours of free RTX 4090 and 5 hours of free CPU time!
HyperAI exclusive invitation link (copy and open in browser):
https://openbayes.com/console/signup?r=Ada0322_NR0n
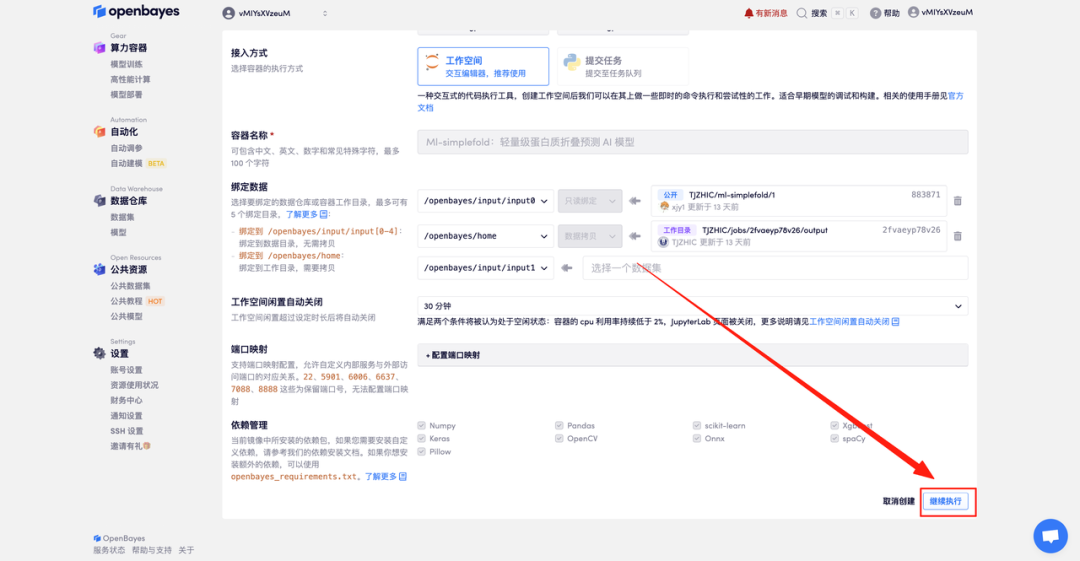
4. Wait for resources to be allocated. The first clone will take about 2 minutes. When the status changes to "Running", click "Open Workspace" to jump to the Demo page.
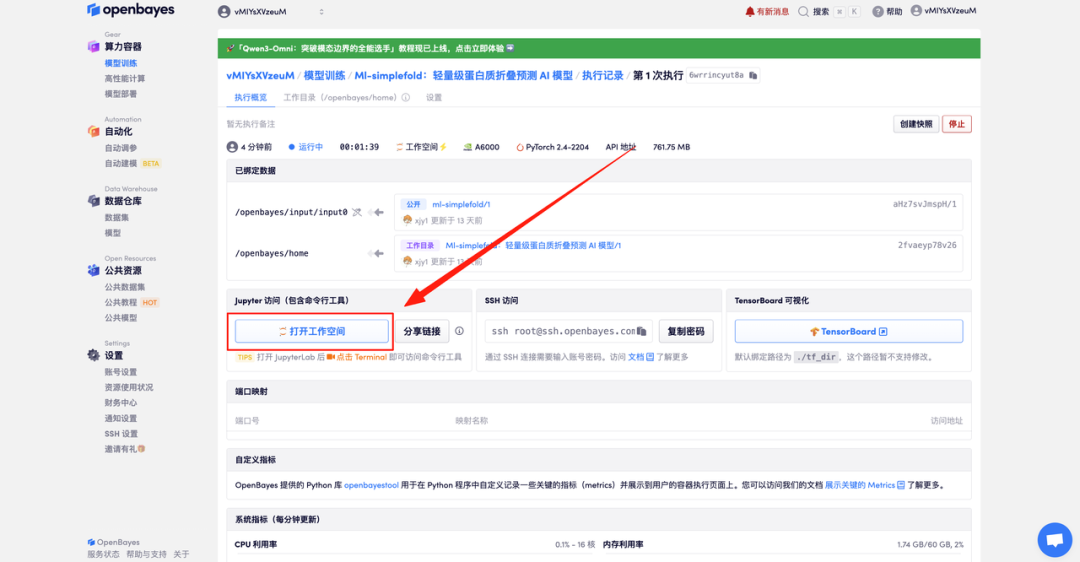
Effect Demonstration
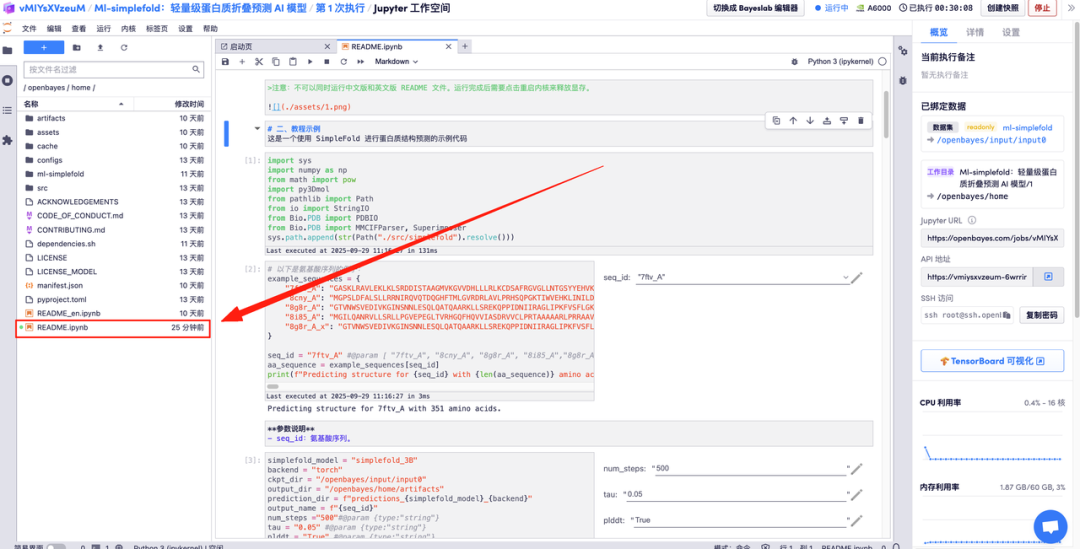
The following is the Ml-simplefold usage page. Click "README" to jump to the generation interface.
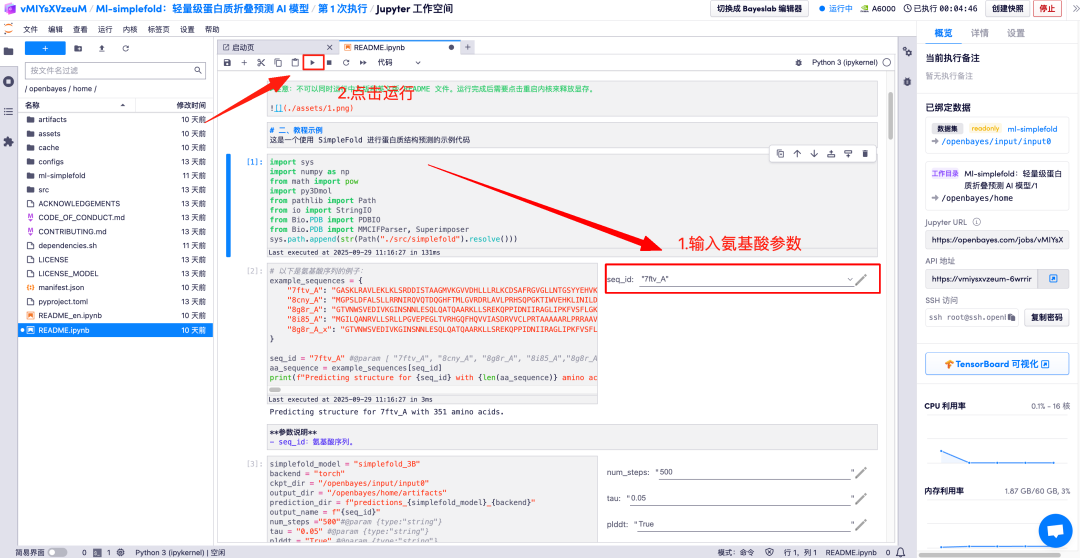
Enter the amino acid parameter descriptions in the corresponding areas and click Run to get the predicted protein file, the full-atom structure, and the 3D visualization image merged with the GT structure.
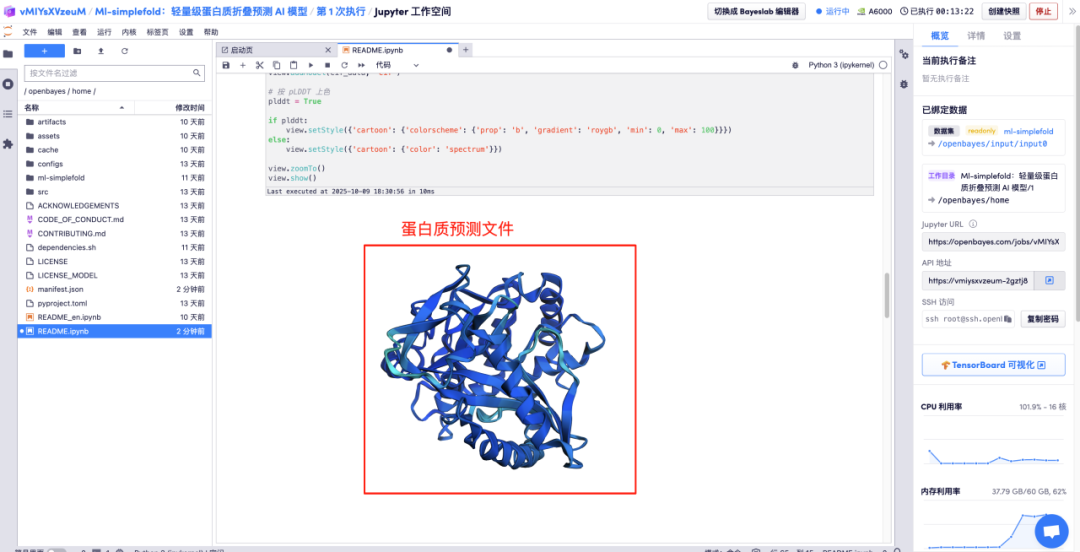
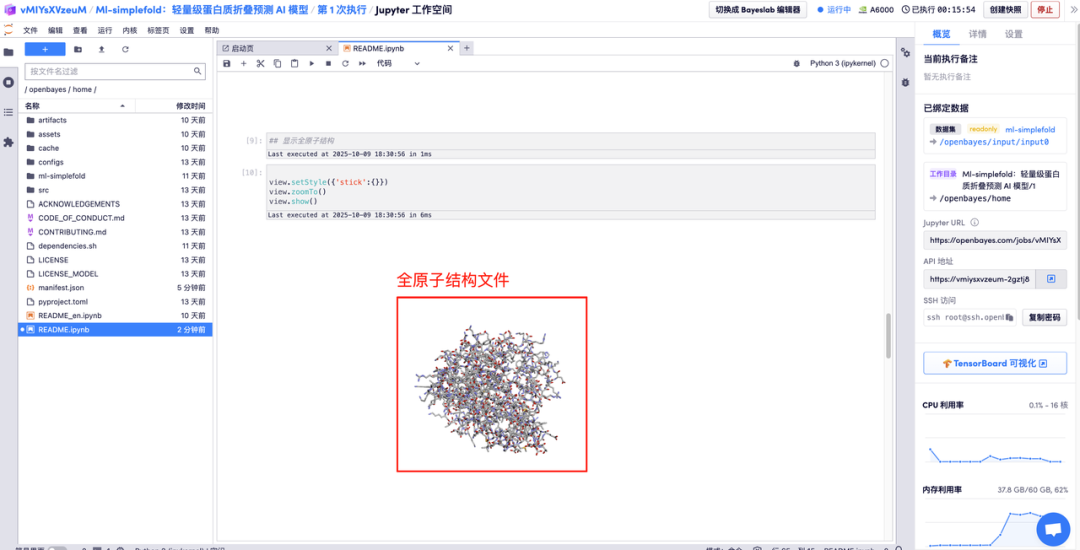
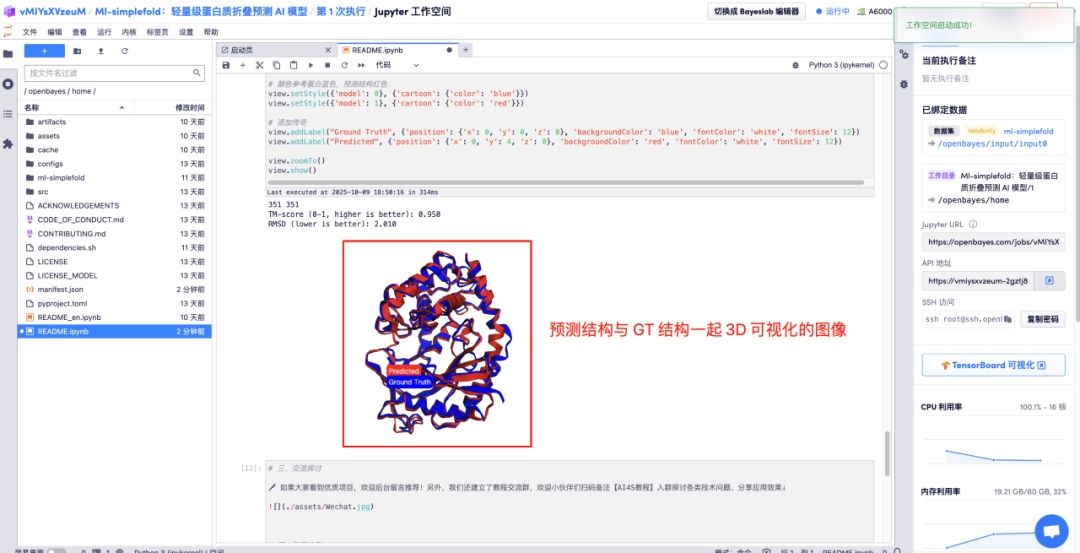
The above is the tutorial recommended by HyperAI this time. Everyone is welcome to come and experience it!
Tutorial Link: