Command Palette
Search for a command to run...
UCLA Releases Bidirectional Brownian Bridge Diffusion Model to Improve the Reproducibility of Virtual Dyeing Results, Significantly Reducing Output Variance
Imaging mass spectrometry (IMS) has been widely used in biology due to its advantages of being label-free, enabling simultaneous multi-molecule imaging, and combining spatial localization with quantification. Compared to traditional mass spectrometry, IMS can map molecular species in biological tissues in a non-targeted, highly multiplexed manner, visualizing the distribution of multiple components at the tissue, cellular, and even subcellular levels. IMS's high spatial resolution demonstrates significant potential in drug target localization, metabolite spatial distribution, and research on drug production mechanisms, advancing the diagnosis and detection of major diseases such as metastatic cancer, Alzheimer's disease, and Parkinson's disease.
IMS has broad molecular coverage and high chemical specificity, butMost IMS platforms lack microscopy-level spatial resolution and cellular morphology contrast, making it difficult to directly link molecular profiles to precise cellular features without additional information.To address this limitation, researchers proposed a multimodal method that combines IMS data with optical microscopy and developed an IMS-microscopy integration method to enhance the interpretability of IMS data through spatial sharpening of IMS data, prediction of out-of-sample molecular distribution, and mining of IMS-derived molecular spectra based on microscopy features.
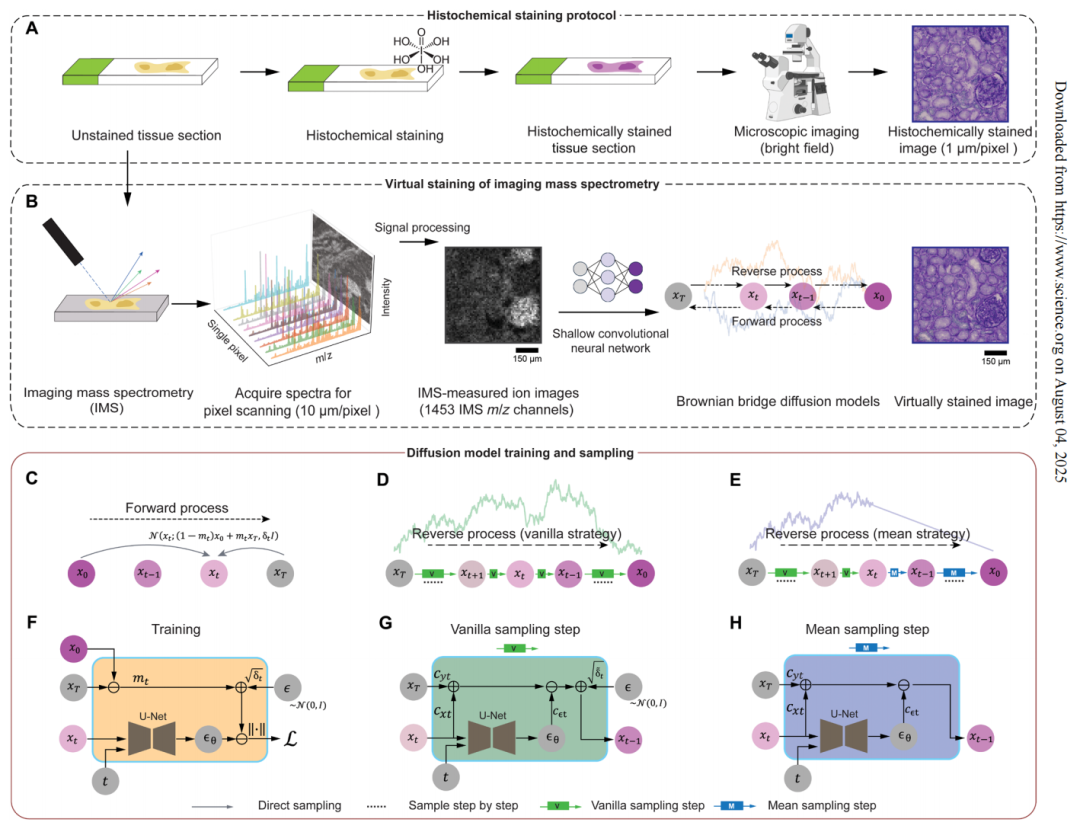
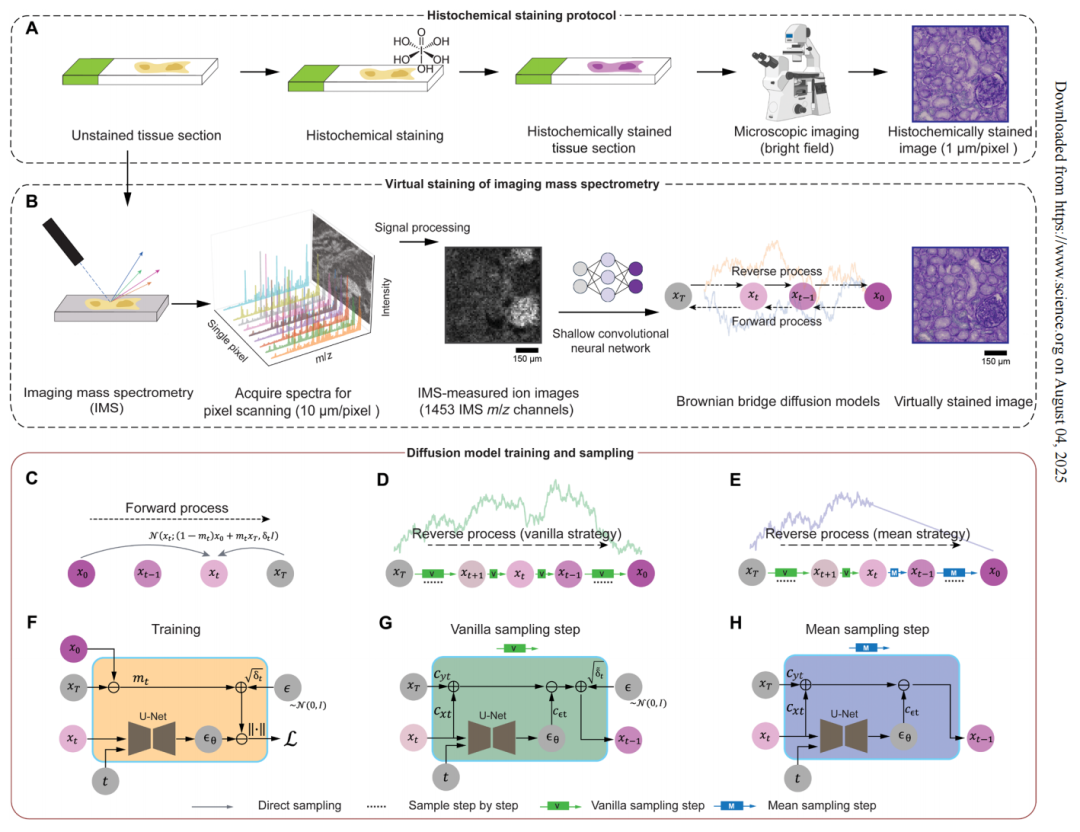
However, the reliance of multimodal methods on bright-field images of histochemical staining or immunofluorescence images increases the complexity of the experiment.A research team from UCLA proposed a virtual histological staining method based on a diffusion model to enhance spatial resolution and digitally introduce cell morphology contrast into mass spectrometry images of label-free human tissues. This enables the prediction of high-resolution cell tissue pathological structures based on low-resolution IMS data, simplifying the workflow of IMS multimodal methods in molecular histology.In addition, the study also used optimized noise sampling during the inference process of the diffusion model to achieve repeatable virtual coloring.
The related research results were published in Science Advances under the title "Virtual staining of label-free tissue in imaging mass spectrometry."
Research highlights:
* The Brownian Bridge Diffusion Model (BBDM) was first applied to IMS data, enabling the conversion from low-resolution ion images of unlabeled tissue to high-resolution virtual staining images;
* Optimize the noise sampling strategy and introduce a deterministic mean sampling strategy in the back diffusion process;
* Proposed a channel selection strategy based on signal-to-noise ratio (SNR), verifying the redundancy and efficient utilization of IMS molecular information.
Paper address:
Follow the official account and reply "Imaging Mass Spectrometry Staining" to get the full PDF
More AI frontier papers:
Label-free tissue mass spectrometry dataset preprocessing
The dataset used in this study includes imaging mass spectrometry ion images (Ion Images) of unlabeled human kidney tissue and high-resolution bright-field images (Bright-field Images) of the same tissue samples. The IMS data for each tissue sample were acquired using pixel-level raster scanning with a lateral spacing of 10 μm, generating a total of 1,453 single ion images containing mass-to-charge ratio channels.
After acquiring IMS data by pixel-level raster scanning, the bright field image of the high-resolution tissue sample is registered with the corresponding IMS ion image.To assist with registration, the research team also acquired autofluorescence microscopy images of tissue sections before and after IMS data acquisition.
This study divides the preprocessing of raw IMS data into four steps:
* Export raw data to a custom binary format and reconstruct it into a pseudo-contour mass spectrum;
* Correct spectrum misalignment based on internal identification peak alignment data;
* Calibrate the dataset mass axis using the theoretical masses of internally identified peaks;
* Normalization correction factors were calculated to normalize mass spectra and ion images using the total ion current method and to calculate the average mass spectrum for each data set based on all pixels.
The dataset used in the experiment contains tissue sample image data from 14 patients. Among them, 712 pairs of enhanced IMS-PAS microscopic image blocks from 4 patients are used for model training, and 201 pairs of unenhanced IMS-PAS microscopic images from the remaining patients are used for model testing.
Virtual histological staining based on bidirectional Brownian bridge diffusion model
In this study, the team converted label-free imaging mass spectrometry data into virtual staining images based on the core generation framework of the Brownian Bridge Diffusion Model (BBDM). Different from the traditional Brownian Bridge design,The diffusion model adopts a bidirectional Brownian bridge design to ensure that its forward process is opposite to the classical denoising diffusion probability model, and realizes the degradation from the target high-resolution bright field image to the low-resolution IMS input image.The reverse process can directly reconstruct the target image based on the IMS input data.
also,The Brownian bridge diffusion model adopts a deterministic mean sampling strategy in the sampling stage, which improves the consistency and repeatability of the generated images.After the strategy reaches a specific time point, the posterior noise introduced during the standard random sampling process is eliminated. Compared to models that end with pure noise and rely entirely on conditional signals, the Brownian Bridge Diffusion Model achieves a more stable image transformation. The entire inference process only requires low-resolution IMS data as input, which the model gradually removes and generates a high-resolution virtual stained image.
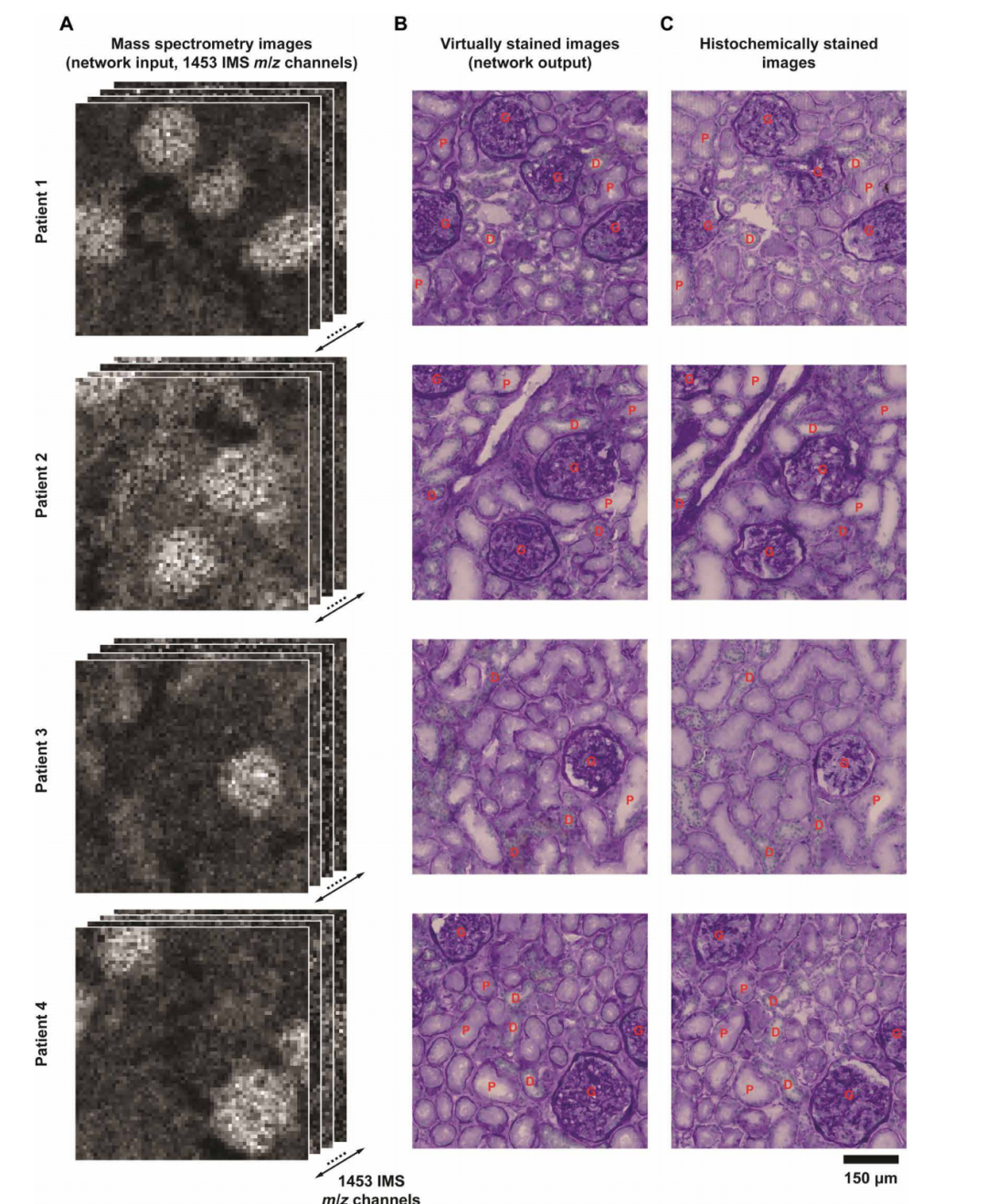
After training, the study evaluated the Brownian bridge model using an independent human kidney tissue test dataset that had not been trained and validated.The virtual staining images generated by the diffusion model based on lower-resolution IMS data show a high degree of visual consistency with the real bright-field staining images.
At the same time, the key kidney structures also showed highly similar results in the virtual stained images and the real stained images.The robustness and good generalization ability of the Brownian bridge framework in virtually coloring low-resolution IMS data are fully verified.
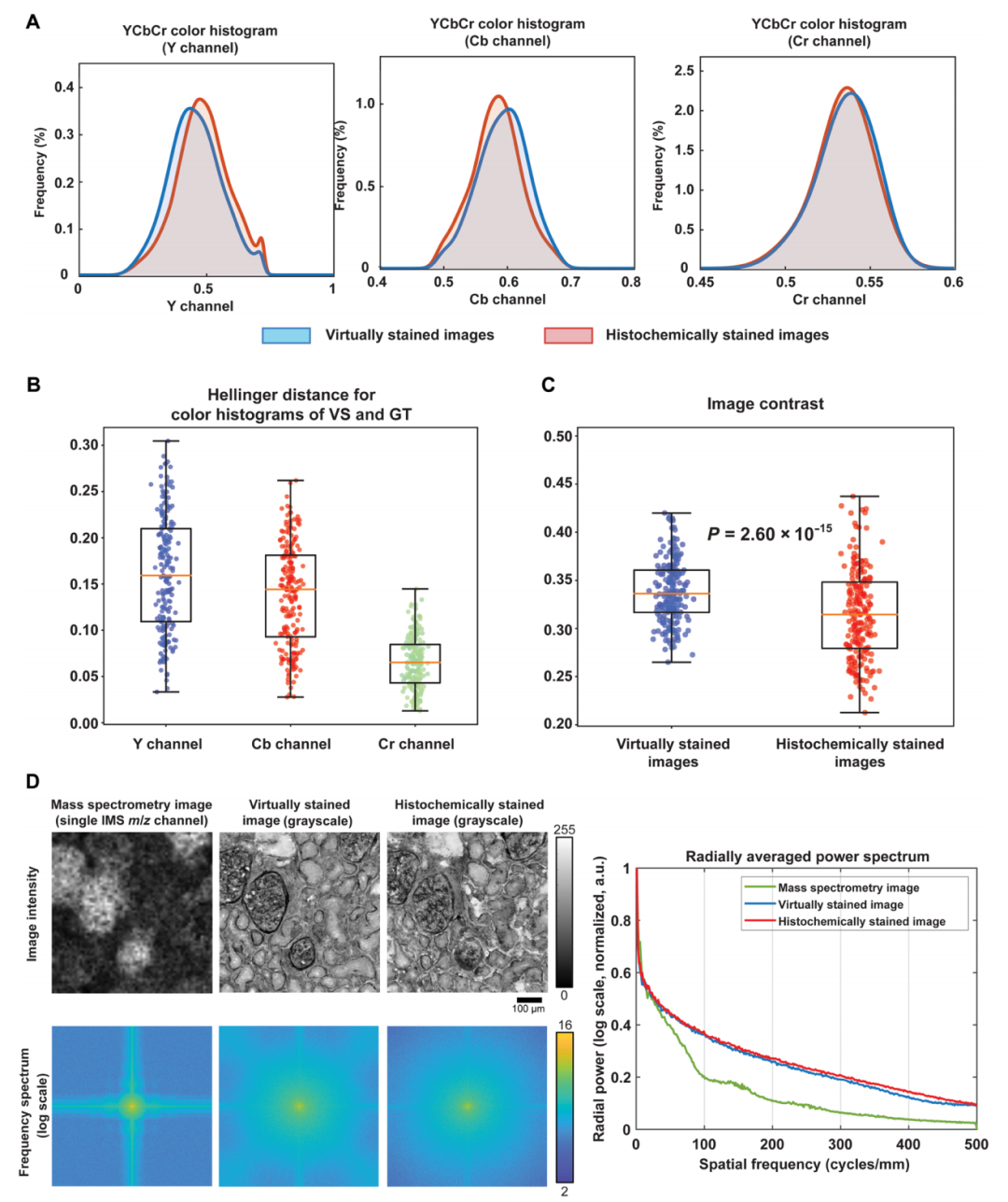
Quantitative evaluation of virtual staining model performance
The experiment confirmed that the virtual staining model has significant super-resolution capabilities through spatial spectrum and color distribution analysis.Spatial spectrum analysis reveals that compared to the input low-resolution mass spectrometry image, the high-resolution image exhibits a relatively large increase in spatial spectrum across all frequency ranges, achieving a substantial improvement in resolution. Furthermore, the radial mean power spectrum of the virtual stained image closely matches that of the high-resolution real stained image, accurately reproducing the spatial frequency characteristics of biological structures.
At the same time, comparative experiments on unpaired high-resolution images further verified the specificity of the virtual staining model.Experiments show that the spectral characteristics of unpaired samples are significantly different from those of paired results, and the color histogram similarity of paired low-resolution and high-resolution images is significantly better than that of unpaired samples, verifying the model's ability to reproduce the inherent color characteristics of tissue chemical staining.
Quantitative comparison between virtually stained PAS images generated from label-free IMS data and their HS counterparts
Number of mass spectrometry image channels
To investigate the impact of the number of mass spectrometry imaging channels on the performance of virtual staining models, the study compared staining strategies with varying channel counts. The research team evaluated a series of virtual staining models with varying numbers of IMS channels based on a fixed test dataset. The models had 363, 91, and 23 IMS channels, representing a 4x, 16x, and 64x reduction, respectively, compared to the original 1,453 channels. For each channel reduction, the experiment evaluated three different selection strategies:
* Signal-to-noise ratio priority strategy: Sort the 1,453 channels in descending order based on signal-to-noise ratio and select the channel with the highest ranking;
Signal-to-noise ratio: mean/standard deviation of all pixels in a channel.
* Frequency priority strategy: Perform a two-dimensional Fourier transform on the ion image of each channel, calculate the ratio of the average power of the high-frequency component to the low-frequency component, and select channels in descending order based on this ratio;
* Uniform sampling strategy: Starting from the first channel, channels are selected at fixed intervals, such as every 4th channel for 4x reduction, every 16th channel for 16x reduction, and every 64th channel for 64x reduction. The channel set selected by each strategy remains consistent throughout the training and testing process of its corresponding model.
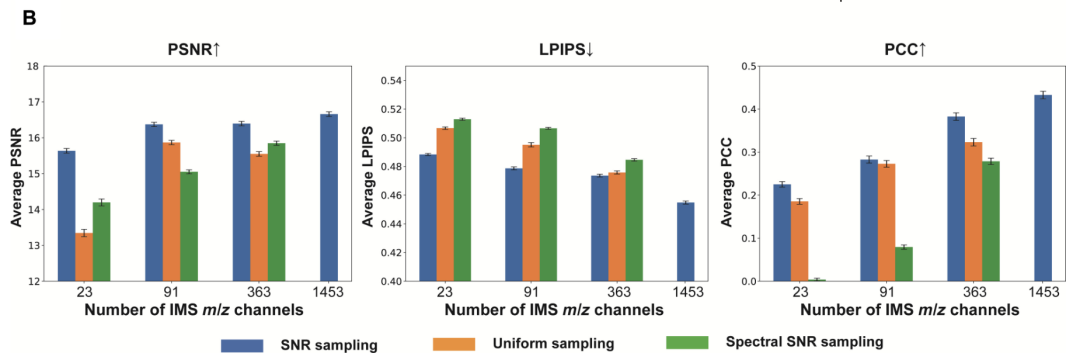
The evaluation results show thatAs the number of IMS channels used decreased from 1,453 to 23, the performance of the virtual staining models under the three strategies declined, and key features such as nuclear morphology were significantly lost.The study used peak signal-to-noise ratio, learned perceptual image patch similarity (LPIPS) and Pearson correlation coefficient to conduct quantitative analysis on the test set and verified thatThe fidelity of the virtual staining model improves significantly with the number of channels.
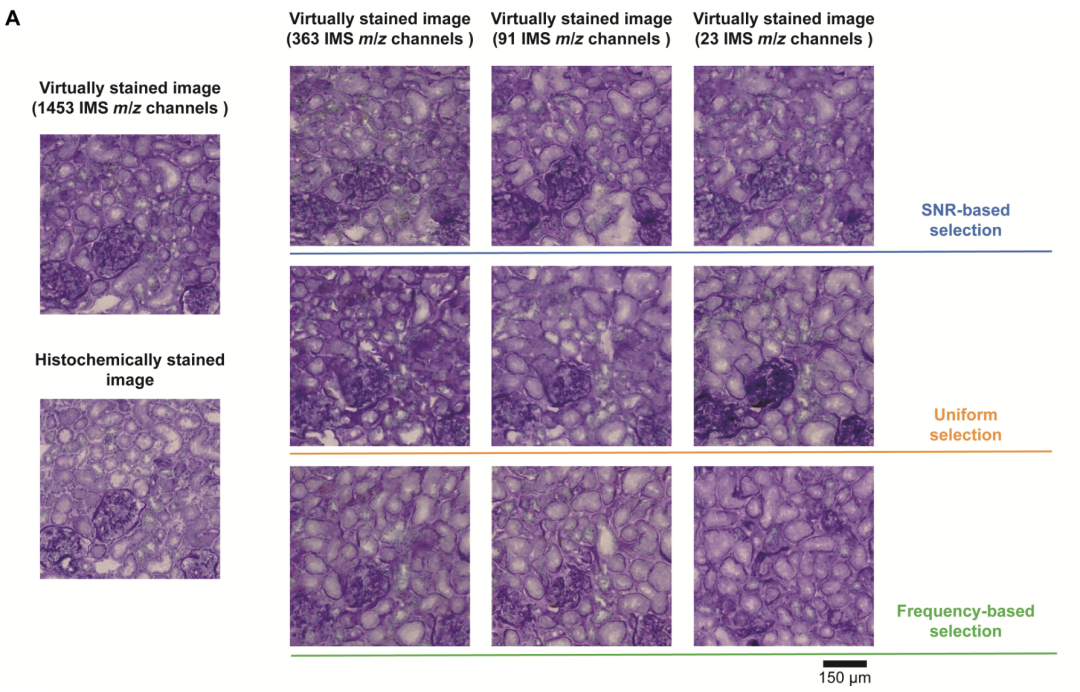
Visual comparison of virtual stained images obtained by three different selection strategies
In addition, among the three channel selection strategies,Compared with the signal-to-noise ratio priority strategy, the frequency priority strategy and the uniform sampling strategy, all of them show significant performance advantages.It was verified that the virtual staining model can more effectively preserve key molecular information when the number of channels is small.
Noise sampling to reduce the output variance of the virtual coloring model
In virtual staining based on diffusion models, the randomness of noise sampling during the reverse diffusion process becomes the main cause of fluctuations in output results. To address the fluctuations in output results, the study proposed a noise sampling engineering strategy that does not require fine-tuning the model. By avoiding the surge in noise variance at the end of reverse diffusion, the consistency of virtual staining results under the same unlabeled tissue field of view is improved. The research team compared three sampling methods based on the trained Brownian bridge model:
* Vanilla sampling: standard reverse diffusion process;
* Mean Sampling: Average the noise path after the Engineered Exit Point;
* Skip Sampling: Predicting the target image domain of the high-resolution brightfield image directly through the denoising network after the project exit point.
The engineering exit point is a critical time threshold artificially set in the reverse generation process of the diffusion model. It is used to switch between random sampling and deterministic calculation strategies in the noise sampling path to suppress random fluctuations in the output results.
The study repeated all three sampling methods five times and evaluated their consistency by calculating the pixel variance coefficients between repeated virtual staining processes. Compared with the original method, both mean sampling and jump sampling strategies effectively reduced the variance in the sampled virtual staining images.
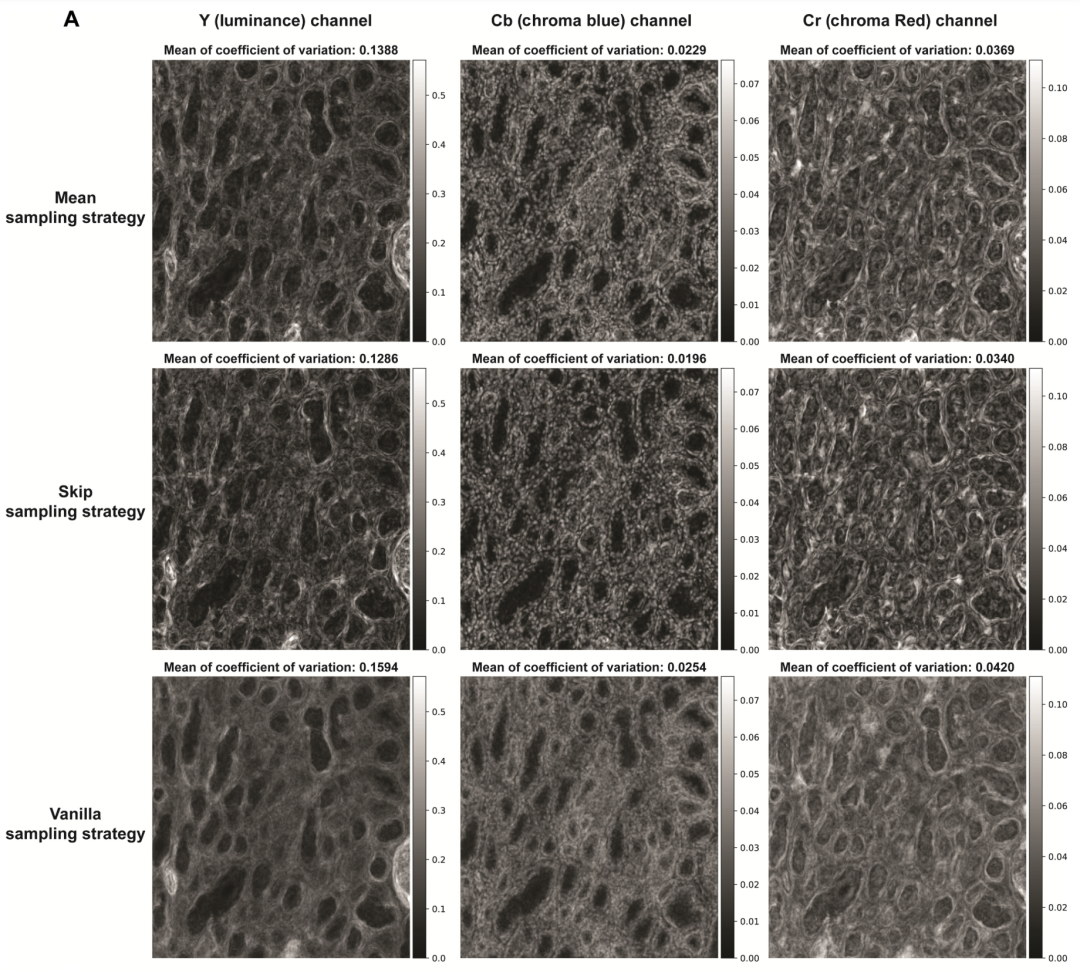
In addition, the researchers calculated the average pixel variance value of all pixels in the unlabeled tissue field of the test image. From the results, compared with the original sampling,Both mean sampling and jump sampling strategies can effectively achieve lower output variance, proving that the diffusion-based virtual dyeing process under this strategy is repeatable.
At the same time, the jump sampling strategy produces lower average coefficient of variation values compared to the mean sampling method.However, the mean sampling strategy produces results that show higher perceptual similarity to the high-resolution images of stained tissue sections, achieving the desired lower average learned perceptual patch similarity.
The study also averaged the results of different inference tests on the same tissue field to further evaluate the performance of the mean sampling strategy. The results showed that the mean sampling strategy only showed stronger performance in scenarios where high consistency was prioritized over image contrast.
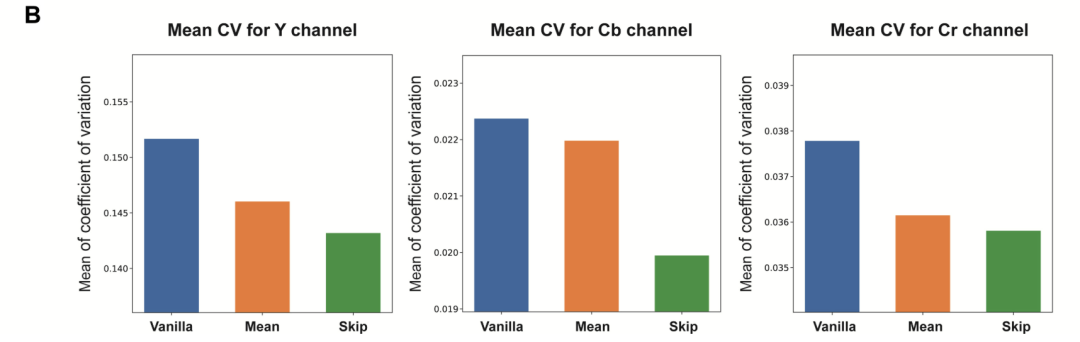
The average coefficient of variation calculated for all test image fields using the three sampling strategies
A new paradigm for virtual staining analysis
Virtual staining techniques based on diffusion models are emerging as a new paradigm for analyzing label-free tissue morphology. Aydogan Ozcan's team at UCLA has previously achieved numerous breakthroughs in integrating diffusion models with tissue staining. In May 2025, the team used the Brownian bridge diffusion model to enhance the spatial resolution and fidelity of label-free virtual tissue staining, performing pixel-by-pixel super-resolution virtual staining of label-free tissues, overcoming the limitations of traditional deep learning-based methods.
The research team integrated sampling technology into the diffusion model-based image inference process, significantly reducing the variance of the generated virtual staining images. When the research team blindly applied the pixel super-resolution virtual staining model to low-resolution autofluorescence images of unlabeled human lung tissue samples, it showed that it consistently outperformed traditional methods in terms of resolution, structural similarity, and perceptual accuracy.And successfully achieved 4 to 5 times pixel super resolution.This research provides a new path with great potential for research in the field of clinical diagnosis.
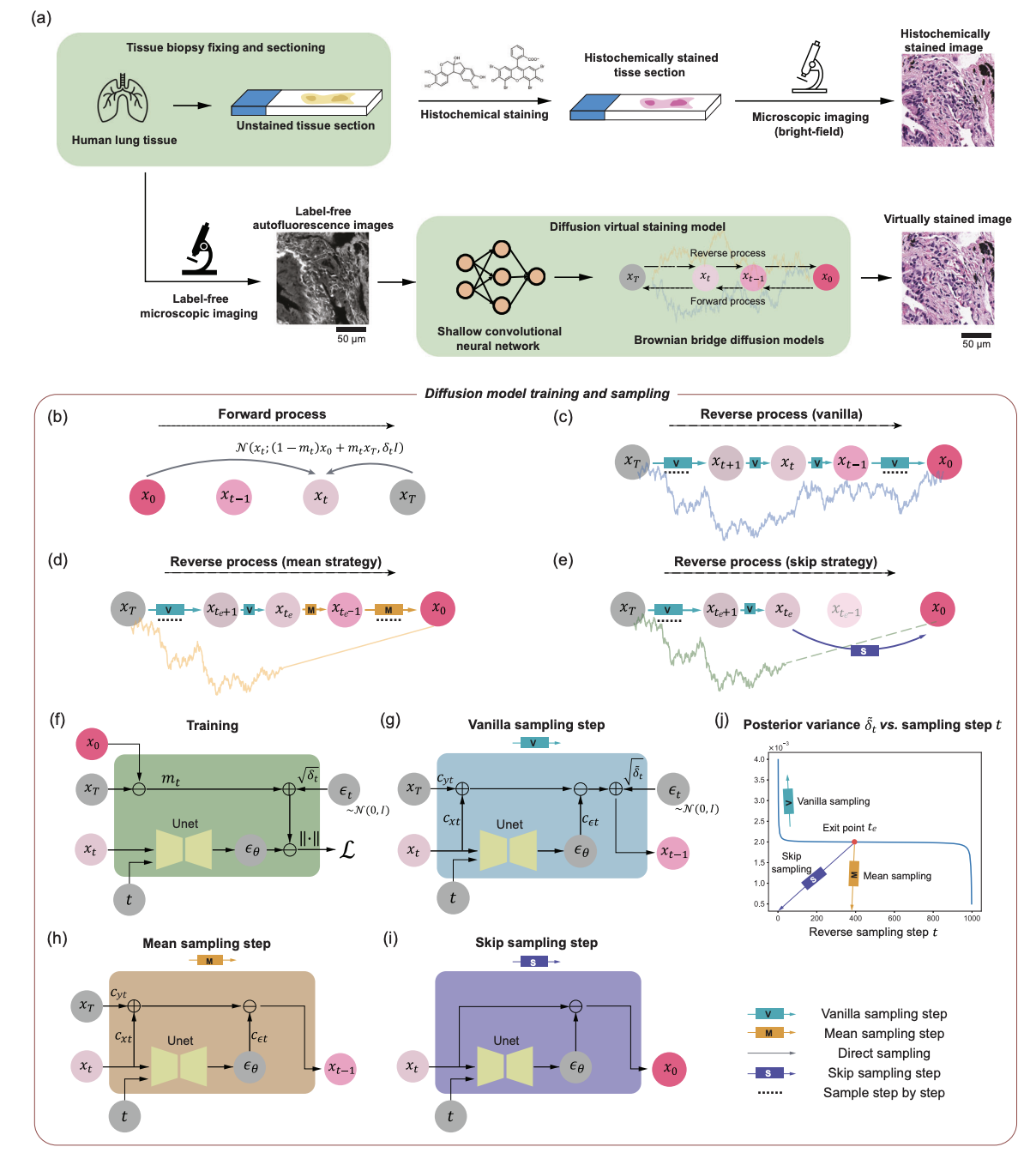
Super-resolution virtual staining of unlabeled tissue sections based on diffusion modeling
The research results were published in Nature Communications under the title "Pixel super-resolved virtual staining of label-free tissue using diffusion models."
Paper address:
https://www.nature.com/articles/s41467-025-60387-z
Continued progress in AI in virtual tissue staining technology has reduced the tedious steps involved in traditional histochemical staining. To address the potential for hallucinations and artifacts in these virtually stained tissue images, Aydogan Ozcan's team has proposed an autonomous quality and hallucination assessment model (AQuA) for virtual tissue staining and digital pathology.
AQuA is able to detect the true value of histochemical staining without the need to obtain the true value of histochemical staining.The system achieved an autonomous accuracy of 99.81 TP3T in detecting acceptable and unacceptable virtual stained tissue images, and achieved a 98.51 TP3T agreement with manual evaluation results by certified pathologists.It demonstrates broad adaptability across a variety of virtual and histochemically stained human tissue images.
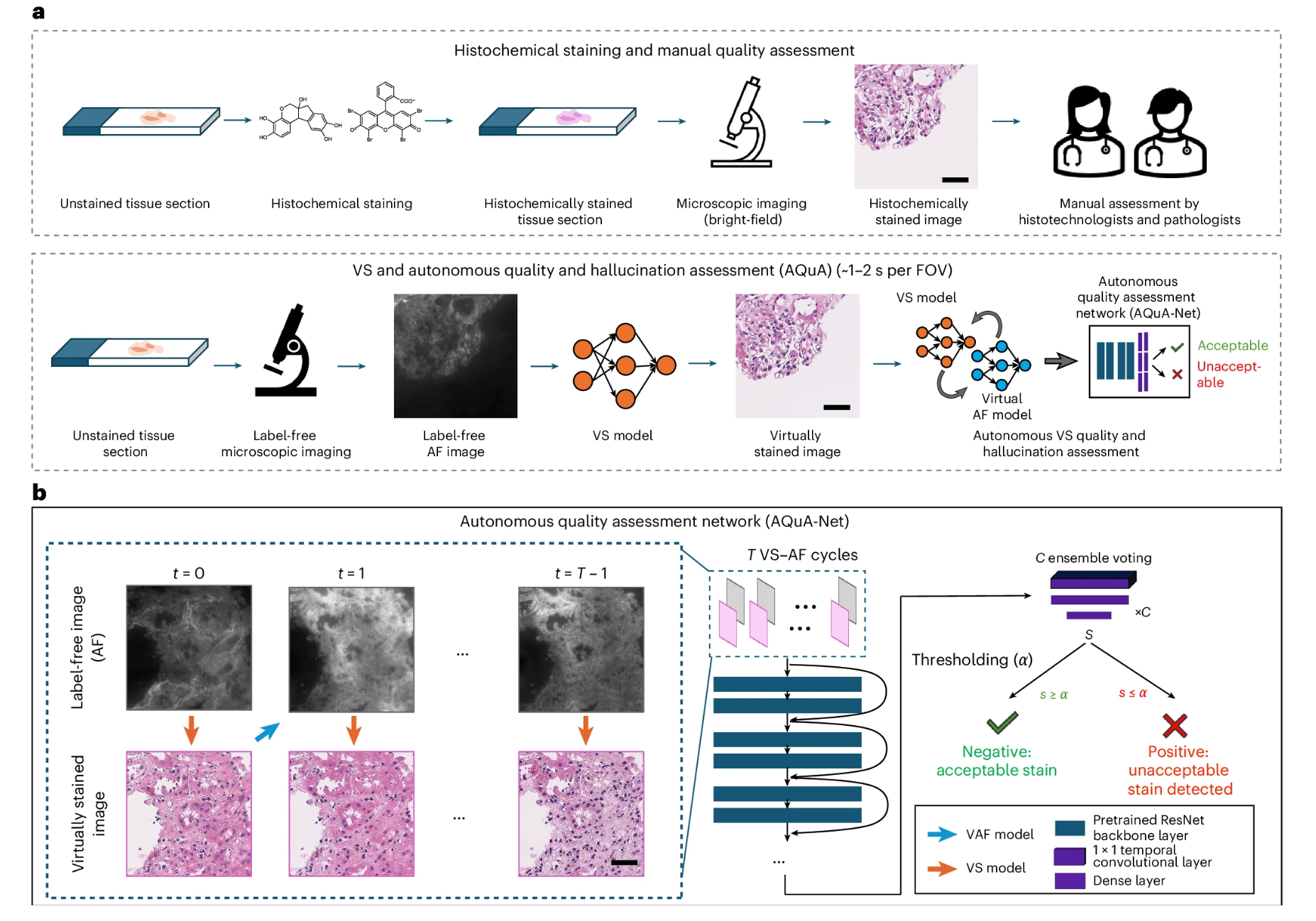
This framework enhances the reliability of virtual tissue staining and provides autonomous quality assurance for image generation and conversion tasks in digital pathology and computational imaging. The research findings, titled "A robust and scalable framework for hallucination detection in virtual tissue staining and digital pathology," were published in Nature Biomedical Engineering.
Paper address:
https://www.nature.com/articles/s41551-025-01421-9
Beyond virtual staining technology, AI applications in clinical diagnosis are expanding at an unprecedented scale and depth. AI models' efficient integration of vast amounts of medical data is driving more accurate early risk prediction for major diseases like cancer. While improving clinical efficiency, AI is also gradually transforming traditional medical assessment, decision-making, and treatment models.