Command Palette
Search for a command to run...
Joint Training Is Possible Without Sharing Data! The UCL Team Is Reshaping Blood Morphology Examination Using Federated learning.
Blood morphology examination is a crucial step in the clinical diagnosis of blood disorders. By observing cell morphology in peripheral blood smears (PBS) or bone marrow aspiration (BMA), physicians can determine the type of leukemia, anemia, infection, and hereditary blood disorders. However, this process is not only labor-intensive but also highly dependent on experienced professionals. Especially in low- and middle-income countries (LMICs), skilled specialists are scarce, making rapid, reliable, and scalable hematological diagnostics a pressing issue.
In recent years, the development of artificial intelligence and deep learning has provided new solutions for blood morphology analysis. AI models can automatically identify different types of white blood cells and assist doctors in making rapid diagnoses.Research indicates that deep learning has significant potential in automated hematological diagnostics.However, significant challenges remain in real-world applications—model training is highly dependent on data, while clinical data is typically distributed across different hospitals and suffers from variations in staining methods, imaging equipment, and a few rare cell types. This data heterogeneity can lead to decreased generalization ability of the model in new institutions or patient populations.
More importantly, medical data involves patient privacy, and cross-institutional data sharing is strictly limited. Traditional centralized training methods typically require the aggregation of large amounts of sensitive medical data and rely on high-performance computing resources, which is difficult to implement in many institutions. How to achieve multi-institutional collaborative training while protecting privacy has become a key issue that urgently needs to be addressed in the field of medical AI.
In this context,A research team from the Department of Computer Science at University College London (UCL) has proposed a federated learning framework for white blood cell morphology analysis.This enables institutions to conduct collaborative training without exchanging training data. Utilizing blood smears from multiple clinical sites, the federated model learns robust and domain-invariant feature representations while maintaining complete data privacy. Evaluations on convolutional networks and Transformer-based architectures demonstrate that federated training outperforms centralized training in cross-site performance and generalization to unknown institutions.
The related research findings, titled "MORPHFED: Federated Learning for Cross-institutional Blood Morphology Analysis," have been published as a preprint on arXiv.
Research highlights:
* Compared to centralized training, federated training demonstrates superior performance across sites and its ability to generalize to unknown institutions.
This method enables collaborative model training across institutions without sharing raw data, providing a viable solution for resource-constrained healthcare environments.

Paper address:
https://arxiv.org/abs/2601.04121
Follow our official WeChat account and reply "MORPHFED" in the background to get the full PDF.
Dataset: Reflecting heterogeneity in real-world clinical settings
This study used blood smear data from multiple medical institutions to ensure that the training data not only covered different cell types but also reflected the heterogeneity in real-world clinical settings.
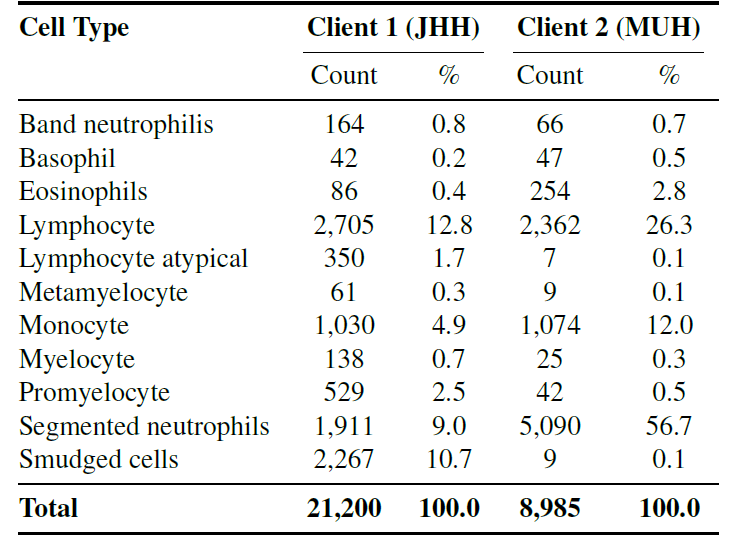
Specifically, the study used independent datasets from two centers.These two datasets contain 11 common cell types (such as neutrophils, eosinophils, basophils, promyelocytes, etc.).It ensures consistency in classification objectives while preserving differences in staining and imaging, and is used to test the generalization ability of federated learning in real heterogeneous environments.
The following figure shows the distribution of different client categories.
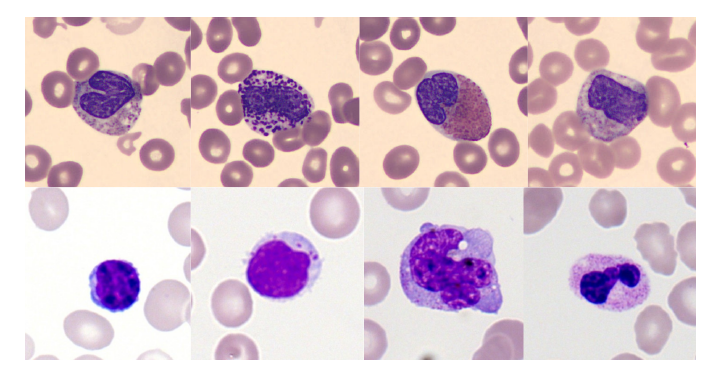
The image below shows examples of some cell types from two training datasets.The difference in coloring style is clearly observable, which is precisely the data bias that the model needs to overcome.
In addition, in order to independently evaluate the model's performance on completely unseen institutional data,The study preserved 12,992 images from the Barcelona Clinical Hospital (Client 3).This dataset serves as an external validation set. It includes various imaging devices, staining methods, and patient populations, and is used to test the model's generalization ability in real-world, cross-institutional scenarios.
Two types of deep learning architectures and four federated aggregation strategies
This study employs two types of deep learning architectures:
* ResNet-34: A classic architecture based on convolutional neural networks (CNNs), using ImageNet pre-trained weights.
* DINOv2-Small: Based on the self-supervised Vision Transformer (ViT), it captures global image features through self-supervised learning.
Training follows a unified protocol: the federated model performs 5 rounds of global communication, with each client performing 5 local training cycles per round, for a total of 25 training cycles; the centralized baseline model uses 25 training cycles and performs 4-fold cross-validation, as shown in the figure below.The data was divided into a 60% training set, a 13.33% validation set, a 13.33% local test set, and a 13.33% global test set.All images were resized to 224×224 pixels and a conservative data augmentation strategy (translation ±10%, rotation ±5°) was employed to preserve diagnostic morphological information.
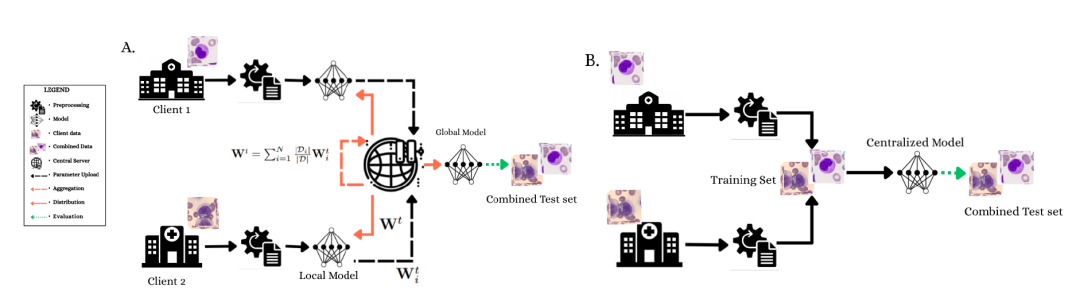
(A) The federated learning framework demonstrates a privacy-preserving collaborative training process in which Client 1 and Client 2 train the model locally and the parameters are aggregated on a central server.
(B) Centralized training paradigm with full access to merged datasets and 4-fold cross-validation.
Both architectures employed selective fine-tuning: ResNet-34 froze the early layers and trained only the last three residual blocks (approximately 11M parameters); DINOv2-Small froze the first 8 Transformer blocks (0-7) and trained blocks 8 through 11 (approximately 9M parameters). Client 3 data was kept isolated throughout the training process and used only to evaluate the final model's generalization ability to new institutional data.
In the federated learning framework, the central server is responsible for coordinating training and distributing global parameters, but does not access the original data; the client trains locally and only returns parameter updates.
The study employed four federated aggregation strategies:
* FedAvg: Calculates a weighted average of client parameters, which is sensitive to extreme class distributions.
* FedMedian: Takes the median value for each coordinate. It is robust to abnormal clients and Byzantine errors, but may suppress minority class signals.
* FedProx: Adds proximal constraints to the local objective function to enhance convergence stability on non-IID data.
* FedOpt: Uses adaptive optimization (Adam) on aggregated gradients to dynamically adjust the learning rate to cope with client heterogeneity and speed up convergence.
Furthermore, to address the severe class imbalance problem, the study combines Focal Loss, weighted random sampling, and gradient accumulation strategies to ensure that training signals from minority classes are not ignored. Gradient clipping (maximum norm 1.0) ensures stable convergence during training.
Model performance was evaluated using balanced accuracy, with a focus on cross-institutional generalization ability to test the model’s robustness when faced with data from different imaging protocols and patient populations.
Federated training demonstrates excellent performance across sites and its ability to generalize to unknown institutions.
To verify the effectiveness of the federated learning framework, the researchers conducted joint test set evaluation and external distributed data generalization evaluation.
① Joint test set evaluation
The model was evaluated on a joint dataset containing data from two clients, and the results are shown in the table below. Different aggregation methods show significant differences in performance across different architectures.
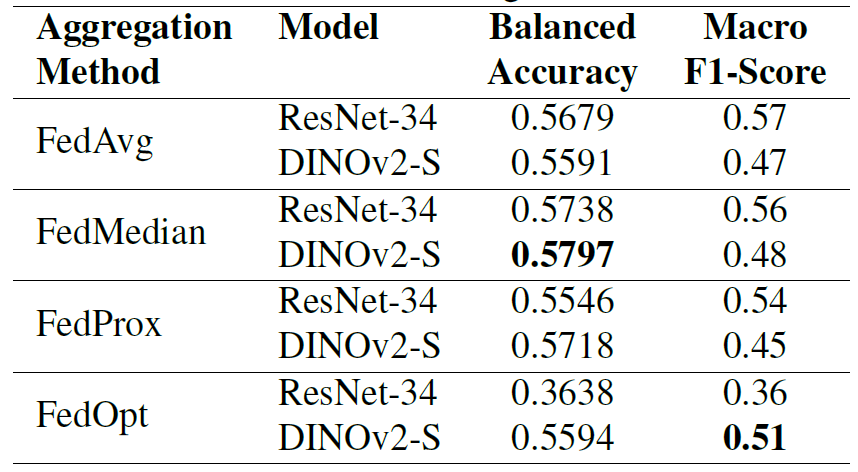
It is worth noting that FedOpt exhibits extreme volatility: it performs very poorly on ResNet-34 (balanced accuracy of 0.3638), while maintaining competitive performance on DINOv2-S (balanced accuracy of 0.5594).In comparison, FedAvg and FedProx showed relatively stable performance on both models;FedMedian performed most consistently across the two architectures, achieving a balanced accuracy of 0.5738 for ResNet-34 and 0.5797 for DINOv2-S.
The results show that federated learning significantly improves performance, demonstrating the advantages of collaborative training without sharing data compared to models trained using only data from a single institution (58% vs 52%, balanced accuracy). Although the federated models perform slightly worse than those trained on all data centrally, they still achieve comparable accuracy while maintaining complete data privacy.
② Generalization evaluation of externally distributed data
Evaluations on the Client 3 external validation dataset from Barcelona show that both federated methods (FedMedian and FedOpt) outperform centralized training on completely unseen institutional data (balanced accuracy 67% vs 64%), as shown in the table below. This indicates that...Exposure to heterogeneous institutional features (such as imaging equipment, patient populations, and staining methods) during federated training helps the model learn more generalizable morphological features.
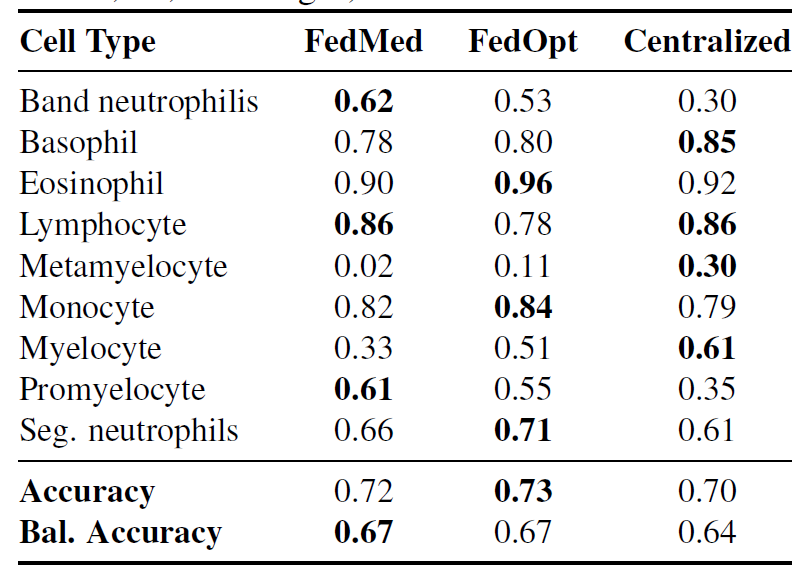
FedMedian showed particularly significant improvements in a minority of cell types: band neutrophils F1: 0.62 vs. central neutrophils 0.30 (an increase of 1071 TP3T), and promyelocytes F1: 0.61 vs. central neutrophils 0.35 (an increase of 741 TP3T).The results show that diagnostic features were effectively preserved under different institutional protocols.However, identification of metamyelocytes remains challenging for all methods (F1: 0.02–0.30), reflecting the fundamental difficulty of learning robust representations from extremely rare classes.
③ The interaction law between architecture and aggregation strategy
The researchers further identified key architecture-aggregation strategy interactions: FedMedian provides cross-architecture robustness but is detrimental to rare classes; FedOpt performs better in preserving cell signal fidelity in minority classes but is architecture-sensitive. The pre-trained Transformer architecture of DINOv2-S exhibits higher robustness to non-IID data distributions, while ResNet-34 is more sensitive to gradient conflicts.
Overall, these findings position federated learning as a robust, privacy-preserving, and generalizable framework for hematological image analysis.
Federated learning becomes key to breaking down healthcare "data silos".
Federated learning is a collaborative machine learning paradigm for distributed data environments. Its core concept is to jointly train models without centralizing the original data. In a federated learning framework, participating institutions (such as hospitals, laboratories, or research centers) train their models locally, uploading only model parameters or gradient updates to a central server. The server then aggregates these updates, generates a global model, and distributes it to each node for further iterative training. Through this mechanism of "data remaining within its domain and models collaborating,"Federated learning enables cross-institutional knowledge sharing while effectively protecting data privacy and meeting stringent data compliance requirements.
In the past few years, many organizations have been working on how to empower the healthcare industry with federated learning. A typical example is Owkin, an end-to-end AI biotechnology company that has been recognized as one of the 20 noteworthy AI startups in France, one of the most noteworthy medical and technology startups in 2023, the Best Medical Technology Award, and one of Forbes AI 50.
Owkin is working on enabling AI technology to identify different biomarkers in multimodal patient data, classify patients into subgroups, match each patient with the best treatment target, promote targeted drug development, optimize disease diagnostic tools, and achieve truly personalized medicine.The key to achieving the above goals lies in how to share data while ensuring patient data privacy?To address this issue, Owkin employs federated learning. To promote the adoption of this technology, Owkin open-sourced its federated learning software, Substra, which can be used in clinical research, drug development, and other applications.
Open source address:
In the field of medical imaging, federated learning is also considered a key technological approach to overcome the challenges of "data silos" and privacy compliance. Medical imaging data is highly sensitive, involving patient privacy and stringent regulations (such as GDPR and HIPAA). Traditional centralized training often faces practical obstacles such as ethical approvals, legal risks, and restrictions on cross-border data transfer. Federated learning enables different hospitals to jointly train models without sharing raw imaging data, thereby improving the model's generalization ability across different devices, staining protocols, and patient populations.Existing research has shown that federated learning can achieve cross-institutional generalization performance close to or even exceeding that of centralized training in fields such as radiological imaging, digital pathology, and ultrasound imaging.It exhibits stronger robustness, especially in external data testing.
From a broader perspective, the "distributed collaborative intelligence" model represented by federated learning is becoming a crucial infrastructure for the large-scale deployment of medical AI in the future. It not only provides a feasible path for training privacy-preserving large-scale medical models but also lays the technological foundation for cross-institutional clinical decision support systems and global collaborative medical research platforms. In specific areas such as blood morphology analysis, federated learning is expected to drive AI from single-institutional laboratory applications to cross-regional and cross-system clinical-grade intelligent diagnostic services, providing key support for precision medicine and digital healthcare.
References:
1.https://arxiv.org/abs/2601.04121
2.https://mp.weixin.qq.com/s/Lf6N7EUHlhibLNc9YXWjTQ