Command Palette
Search for a command to run...
ルーブリック報酬を用いたAI共同研究者の訓練
ルーブリック報酬を用いたAI共同研究者の訓練
概要
AI共同研究者(AI co-scientists)が、人間の研究者が研究目標を達成するのを支援するツールとして登場しつつある。これらのAI共同研究者の重要な特徴の一つは、研究の目的と制約条件をもとに、研究計画を生成する能力である。得られた計画は研究者によるアイデア出しの支援として利用できるほか、さらに精緻化された後には実行可能なものとなる場合もある。しかし、現在の言語モデルは、すべての制約条件および隠れた要件を満たす研究計画を生成する点で依然として課題を抱えている。本研究では、既存の研究論文の膨大なコアスを活用して、より優れた研究計画を生成できる言語モデルを訓練する方法を検討する。複数の分野にわたる論文から自動的に研究目標と、目的に特化した評価基準(グレーディングルーブリック)を抽出することで、スケーラブルかつ多様な訓練データセットを構築した。その後、自己評価(self-grading)を用いた強化学習により、研究計画生成モデルを訓練した。訓練中は初期方策の固定コピーを評価者(grader)として用い、評価基準が生成者と検証者の間のギャップ(generator-verifier gap)を形成することで、外部の人的監督なしに性能向上を実現した。本手法の有効性を検証するため、機械学習分野の研究目標を対象に、専門家による人間評価を225時間にわたり実施した。その結果、微調整済みのQwen3-30B-A3Bモデルが生成した計画は、初期モデルの計画と比較して70%の研究目標で専門家に好まれ、自動抽出された目的別評価基準の84%が専門家によって承認された。汎用性の検証のため、本アプローチを医療分野の論文やarXivの新規プレプリントから得られる研究目標に拡張し、最先端モデルによる審査委員会(jury)を用いて評価した。微調整によって12~22%の相対的な性能向上が得られ、顕著な跨領域一般化能力が確認された。これは、実行フィードバックが得られない医療研究のような課題設定においても、本手法が有効であることを示している。以上の結果から、スケーラブルで自動化された訓練手法が、汎用性の高いAI共同研究者の実現に向けた一歩である可能性が示された。
One-sentence Summary
Meta Superintelligence Labs, ELLIS Institute Tübingen, Max Planck Institute for Intelligent Systems, University of Oxford, and University of Cambridge researchers propose a scalable framework to train AI co-scientists for generating high-quality research plans. They introduce a reinforcement learning approach with self-grading, where a frozen initial model acts as a grader using goal-specific rubrics extracted from scientific papers as privileged information, creating a generator-verifier gap. This method enables improvement without human supervision, validated by human experts who preferred plans from their finetuned Qwen3-30B-A3B model over the initial model for 70% of machine learning research goals, and by strong performance gains across medical and arXiv domains, demonstrating significant cross-domain generalization.
Key Contributions
- We introduce a scalable method to automatically extract research goals and goal-specific grading rubrics from scientific papers across domains, enabling the training of AI co-scientists without relying on costly or infeasible real-world execution feedback.
- By using reinforcement learning with self-grading—where a frozen model acts as a grader with access to privileged rubrics—we close the generator-verifier gap, allowing the plan generator to learn detailed, constraint-aware research strategies without human annotation.
- Human evaluations show a 70% preference for plans generated by our finetuned Qwen3-30B-A3B model over the initial model, and cross-domain tests on medical and arXiv papers demonstrate 12–22% relative improvements, proving strong generalization even in settings where execution feedback is unavailable.
Introduction
The authors address the challenge of training AI co-scientists to generate high-quality, constraint-aware research plans for open-ended scientific goals—critical for accelerating discovery without relying on costly, domain-specific execution environments. Prior approaches either depend on end-to-end simulation (limited to narrow, programmatic tasks) or lack scalable feedback mechanisms, especially in domains like medicine where real-world experimentation is infeasible. To overcome this, the authors propose a scalable, automated training pipeline that extracts research goals and goal-specific grading rubrics from scientific papers across multiple domains. They train a language model via reinforcement learning with self-grading, where a frozen copy of the initial model acts as a grader using the extracted rubrics as privileged information, creating a generator-verifier gap that enables improvement without human annotation. Human evaluations with ML experts show a 70% preference for the finetuned model’s plans, and cross-domain tests on medical and arXiv preprints demonstrate 12–22% relative improvements, highlighting strong generalization and the viability of training generalist AI co-scientists.
Dataset
- The dataset, named ResearchPlanGen, comprises research goals, domain-specific rubrics, and reference solutions extracted from peer-reviewed papers across three domains: machine learning (ResearchPlanGen-ML), arXiv preprints (ResearchPlanGen-ArXiv), and medical research (ResearchPlanGen-Medical).
- ResearchPlanGen-ML includes 6,872 training samples from accepted NeurIPS 2023–2024 and ICLR 2025 papers (via OpenReview), with a test set of 685 samples drawn from top-tier Oral and Spotlight papers from the same conferences.
- ResearchPlanGen-ArXiv consists of 6,573 training samples and 1,496 test samples, stratified across eight quantitative disciplines (Physics, Math, Statistics, Computer Science, Economics, Electrical Engineering, Quantitative Finance, Quantitative Biology), with test data collected from August–September 2025 to prevent model contamination.
- ResearchPlanGen-Medical is sourced from the pmc/openaccess dataset, using peer-reviewed papers published after March 2023; while some overlap with pre-training data is possible, the targeted fine-tuning demonstrates methodological value.
- The authors use the dataset to train and evaluate their model, combining all three subsets with domain-specific mixture ratios during training to ensure balanced representation.
- Data processing includes filtering by publication date, domain relevance, and quality (e.g., excluding low-quality or contaminated samples), with careful curation to maintain representativeness and avoid bias.
- For medical research, the authors apply a strict annotation protocol with anti-LLM safeguards, including redacted prompt injections and enforced policies to prevent automated generation during labeling.
- Metadata construction includes detailed experimental design elements such as sample size, processing timelines, and quality control steps, with full documentation of protocols and controls.
- A cropping strategy is applied in the medical subset by focusing on specific research goals (e.g., sEV DNA analysis in lung cancer) and defining precise experimental workflows, including nuclease digestion steps, spike-in controls, and orthogonal validation methods.
Method
The methodology centers on training a policy model, denoted as πθ, to generate research plans p given an open-ended research goal g. This approach addresses the challenges of acquiring large-scale, high-expertise datasets and establishing automated feedback without relying on expensive human supervision. The framework leverages a language model to extract research goals and goal-specific rubrics from existing scientific papers, which are then used to train the plan generator via reinforcement learning (RL). A key component is the use of a frozen copy of the initial model as a grader, θr(g,p,Rg), which outputs a reward r∈[0,1] based on the generated plan. This allows for iterative training without expert supervision, reserving human evaluation for final assessment.
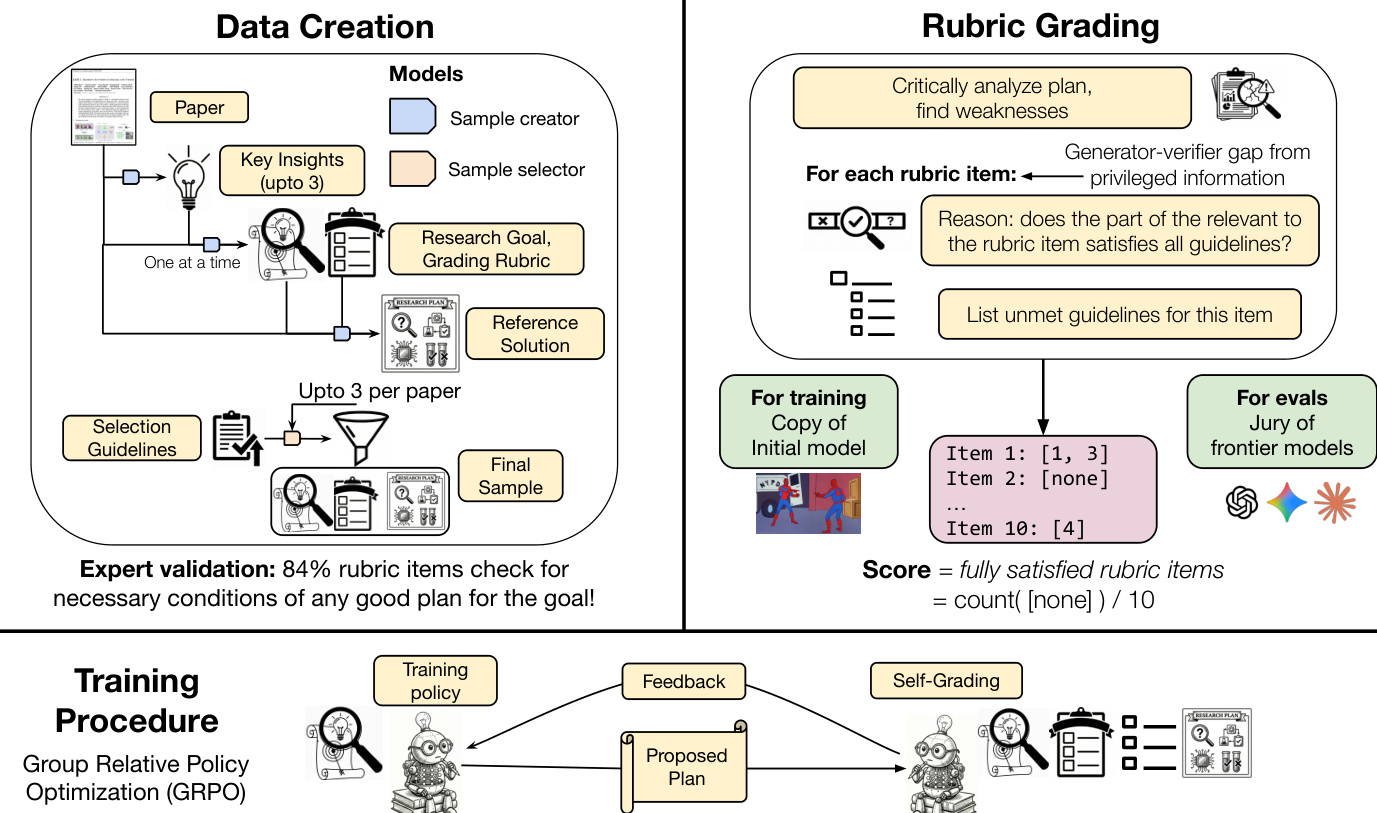
The overall methodology is structured into three main phases: data creation, rubric grading, and the training procedure. The data creation process begins with a sample creator model, which extracts up to three candidate tuples—each containing a research goal, a preliminary set of grading rubrics, and a reference solution—from a single research paper. This process is guided by specific instructions to focus on novel insights. A sample selector model then evaluates each component of the candidate tuples independently. The rubrics are filtered to the top-10 items based on diversity and quality, the goal and rubrics are checked against predefined quality criteria, and the reference solution is graded using the same protocol as the final plan evaluation. The tuple with the highest average score across all components is selected as the final training sample. This automated, scalable, and domain-agnostic process ensures the collection of high-quality training data.
The rubric grading process is designed to provide a reliable and directionally correct reward signal for training. The grader evaluates a proposed plan against two key resources: the goal-specific rubrics and a set of seven general guidelines. For each rubric item, the grader must explicitly identify which, if any, of the general guidelines are violated by the relevant section of the plan. A rubric item is considered satisfied only if the grader returns an empty list of violations. The final score is the fraction of satisfied rubric items. This violation-based grading approach is designed to yield more grounded scores than direct binary or numeric ratings and enables fine-grained error analysis. The general guidelines are derived from common failure modes of language model-generated research ideas and are designed to ensure the plan adheres to fundamental principles of scientific reasoning, such as avoiding vague proposals or overlooked flaws.
The training procedure employs Group Relative Policy Optimization (GRPO) to optimize the policy πθ. This method removes the need for a separate value network by normalizing rewards within a group of outputs for the same input. The primary reward signal is computed by a frozen copy of the initial policy, which acts as the grader. To prevent the model from increasing plan length to maximize the reward, a structural constraint is enforced. The model is permitted unlimited "thinking" tokens to reason through the problem, but the final plan must be enclosed within a strict length limit. Any violation of this constraint results in a penalty. The final numerical reward is calculated as the ratio of satisfied rubric items to the total number of rubric items, minus a penalty for any format violation.
Experiment
- Human evaluation on 100 ML research plans shows the finetuned model is preferred over the initial Qwen-3-30B model in 70.0% ± 5.3% of annotations (p < 0.0001), with average scores of 7.89 ± 0.2 vs. 7.31 ± 0.2, indicating improved plan quality and usefulness for graduate students.
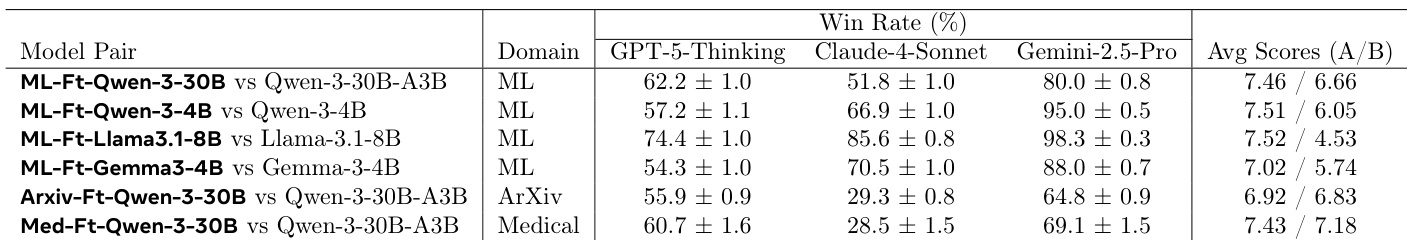
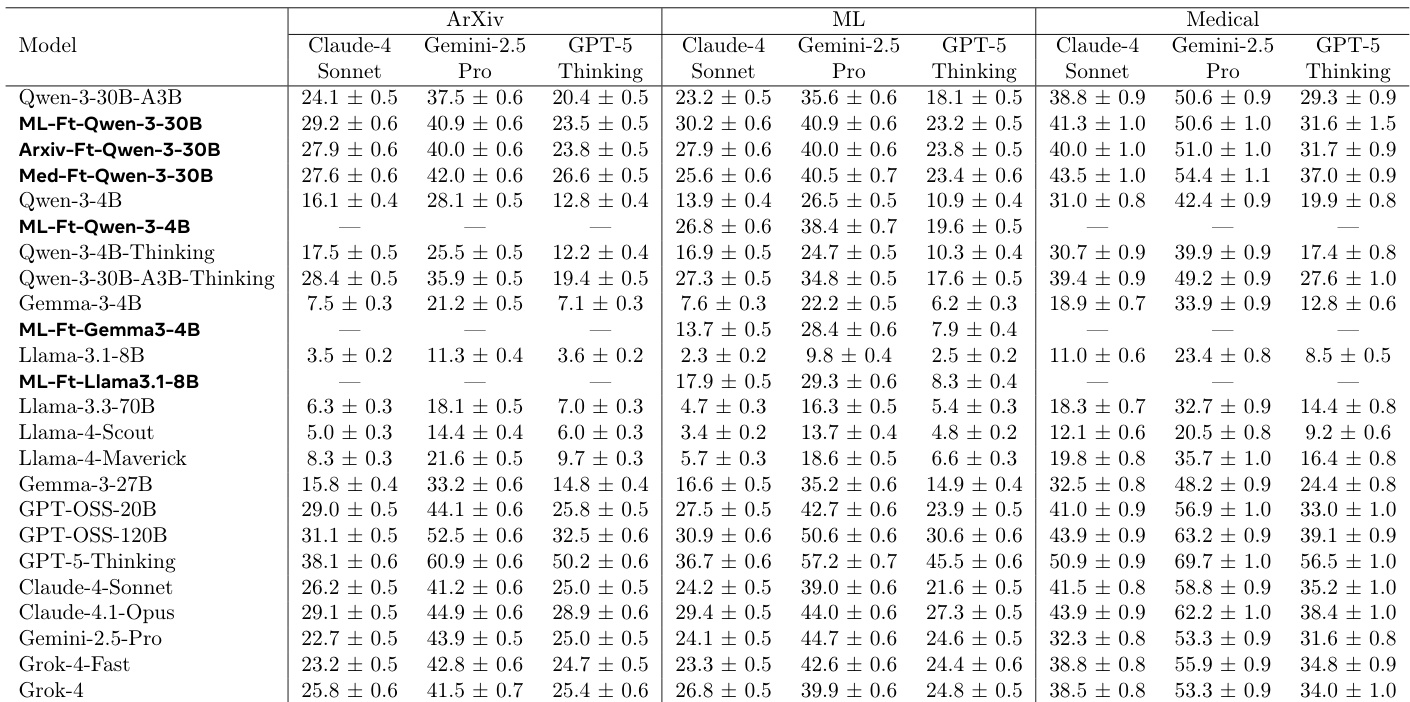
- Automated evaluation across ML, medical, and arXiv domains using a jury of frontier models (GPT-5-Thinking, Gemini-2.5-Pro, Claude-4-Sonnet) shows consistent improvements: finetuned models outperform the initial policy by 10–15% relative gain, with the medical finetune achieving 15% and 17.5% relative improvement on ML and arXiv tasks, respectively.
- Ablation studies confirm that reward modeling with rubrics, disabling the KL penalty, using a stronger 30B MoE reward model, and providing both specific and general guidelines significantly improve performance, while supervised fine-tuning degrades plan quality.
- Cross-domain generalization is demonstrated: domain-specific finetuning (e.g., medical) improves performance on unrelated domains (ML, arXiv), suggesting the model learns general research planning principles.
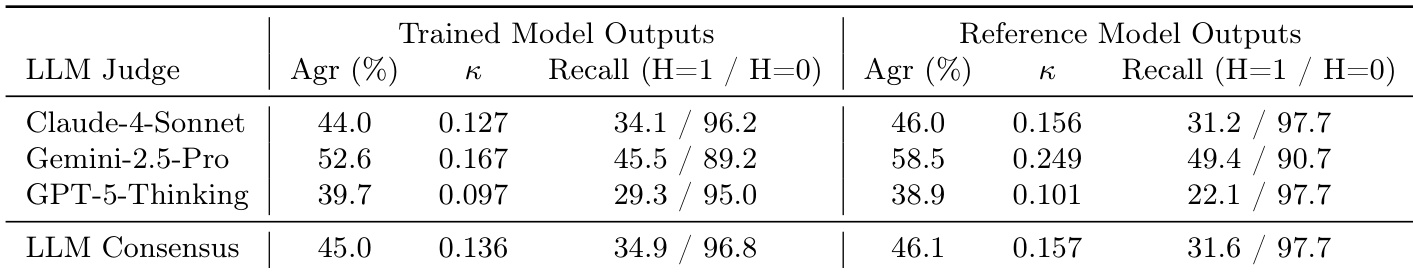
- Human-judge agreement with frontier models is moderate (e.g., GPT-5-Thinking κ = 0.44), and automated grading shows significant signal in aggregate despite per-sample subjectivity, validating its use for scalable evaluation.
The authors use a jury of frontier models to evaluate the performance of finetuned models against their initial counterparts across multiple domains. Results show that the finetuned models consistently outperform the initial models, with win rates ranging from 51.8% to 88.0% depending on the model and domain, and average scores that are higher for the finetuned models in all cases.
The authors compare the performance of trained and reference model outputs using three frontier LLMs as judges. Results show that the trained model outputs achieve higher agreement (Agr) and recall (H=1) across all judges compared to the reference model outputs, with the Gemini-2.5-Pro judge showing the highest performance for both sets. The LLM consensus scores are slightly lower than the individual judges' scores, indicating moderate agreement among the judges.
The authors use human expert annotations to evaluate the quality of research plans generated by a finetuned model compared to an initial model. Results show that the finetuned model's plans are preferred by experts in 70.0% of annotations, with a statistically significant improvement in overall scores (7.89 vs. 7.31 out of 10). The finetuned model also satisfies more rubric items (79.8% vs. 73.8%), indicating higher plan quality across multiple criteria.

The authors use a reinforcement learning framework with self-generated rewards to finetune language models for generating research plans, comparing the initial Qwen-3-30B model to its finetuned variants across ML, ArXiv, and medical domains. Results show that the finetuned models consistently outperform the initial model in automated evaluations, with domain-specific finetuning leading to significant improvements and cross-domain generalization, particularly in the medical domain.
The authors use a validation set and a stronger language model, Claude-4-Sonnet, to determine the optimal stopping step for training, selecting the checkpoint that achieves the best performance on the validation set. The table shows that the stopping step varies by model and finetuning domain, with the Qwen-3-30B model finetuned for ML and ArXiv tasks stopping at step 100, while the PubMed finetune stops at step 140. For the Qwen-3-4B model, the stopping step is 200 for ML finetuning and 100 for the SFT and RL variants, indicating that the optimal training duration depends on the specific model and task.