Command Palette
Search for a command to run...
SkinFlow: Efficient Information Transmission for Open Dermatological Diagnosis via Dynamic Visual Encoding and Staged RL
SkinFlow: Efficient Information Transmission for Open Dermatological Diagnosis via Dynamic Visual Encoding and Staged RL
Lijun Liu Linwei Chen Zhishou Zhang Meng Tian Hengfu Cui Ruiyang Li Zhaocheng Liu Qiang Ju Qianxi Li Hong-Yu Zhou
Abstract
General-purpose Large Vision-Language Models (LVLMs), despite their massive scale, often falter in dermatology due to "diffuse attention" - the inability to disentangle subtle pathological lesions from background noise. In this paper, we challenge the assumption that parameter scaling is the only path to medical precision. We introduce SkinFlow, a framework that treats diagnosis as an optimization of visual information transmission efficiency. Our approach utilizes a Virtual-Width Dynamic Vision Encoder (DVE) to "unfold" complex pathological manifolds without physical parameter expansion, coupled with a two-stage Reinforcement Learning strategy. This strategy sequentially aligns explicit medical descriptions (Stage I) and reconstructs implicit diagnostic textures (Stage II) within a constrained semantic space. Furthermore, we propose a clinically grounded evaluation protocol that prioritizes diagnostic safety and hierarchical relevance over rigid label matching. Empirical results are compelling: our 7B model establishes a new state-of-the-art on the Fitzpatrick17k benchmark, achieving a +12.06% gain in Top-1 accuracy and a +28.57% boost in Top-6 accuracy over the massive general-purpose models (e.g., Qwen3VL-235B and GPT-5.2). These findings demonstrate that optimizing geometric capacity and information flow yields superior diagnostic reasoning compared to raw parameter scaling.
One-sentence Summary
The authors from Baichuan Inc., Peking University First Hospital, Tsinghua University, and the University of Hong Kong propose SkinFlow, a framework that enhances dermatological diagnosis in large vision-language models by optimizing visual information transmission through a Virtual-Width Dynamic Vision Encoder and a two-stage reinforcement learning strategy, achieving state-of-the-art performance on Fitzpatrick17k without relying on massive parameter scaling.
Key Contributions
- Dermatological diagnosis with general-purpose Large Vision-Language Models (LVLMs) is hindered by "diffuse attention," where models fail to disentangle subtle lesions from background noise, leading to inefficient visual information transmission despite massive scale.
- The paper introduces SkinFlow, which optimizes diagnostic reasoning through a Virtual-Width Dynamic Vision Encoder (DVE) that adaptively "unfolds" complex pathological manifolds to enhance signal-to-noise ratio, coupled with a two-stage reinforcement learning strategy that sequentially aligns explicit medical descriptions and reconstructs implicit diagnostic textures.
- On the Fitzpatrick17k benchmark, the 7B-parameter SkinFlow model achieves a +12.06% Top-1 accuracy and +28.57% Top-6 accuracy gain over large general-purpose models like Qwen3VL-235B and GPT-5.2, while a clinically grounded evaluation protocol prioritizes diagnostic safety and hierarchical relevance over rigid label matching.
Introduction
General-purpose Large Vision-Language Models (LVLMs) struggle in dermatology due to "diffuse attention," where they fail to isolate subtle pathological lesions from background noise, limiting diagnostic precision. Existing models also rely on rigid evaluation metrics that ignore clinical relevance, treating all non-exact matches as equally incorrect—despite real-world scenarios where semantically close or therapeutically consistent predictions are clinically valuable. To address these issues, the authors introduce SkinFlow, a framework that reframes dermatological diagnosis as an optimization of visual information transmission efficiency. They propose a Virtual-Width Dynamic Vision Encoder (DVE) that adaptively "unfolds" complex visual manifolds without increasing parameters, significantly improving signal-to-noise ratio. Complementing this, a two-stage reinforcement learning strategy first aligns explicit medical descriptions and then reconstructs implicit diagnostic textures within a constrained semantic space. The authors further establish a clinically grounded evaluation protocol that prioritizes diagnostic safety, hierarchical relevance, and treatment consistency over label matching. Empirically, their 7B model outperforms massive general-purpose models (e.g., Qwen3VL-235B and GPT-5.2) by +12.06% in Top-1 and +28.57% in Top-6 accuracy on the Fitzpatrick17k benchmark, demonstrating that optimizing information flow and geometric capacity yields superior clinical performance compared to raw parameter scaling.
Dataset
- The dataset comprises approximately 5,000 dermatological images sourced from both domestic and international repositories, each labeled with disease category annotations.
- Of these, 4,000 image-caption pairs were generated using a hybrid approach: initial automatic captioning via large language models followed by expert refinement to ensure clinical accuracy and descriptive quality.
- For evaluation, two distinct benchmarks were constructed to reflect real-world diagnostic scenarios without assuming a closed-set disease taxonomy.
- The first benchmark uses 1,000 randomly sampled images from the publicly available Fitzpatrick17k dataset, offering broad coverage of dermatological conditions and serving as a standard benchmark for generalization assessment.
- The second benchmark consists of approximately 200 images curated internally, with all labels independently verified and corrected by board-certified dermatologists from Class-III Grade-A hospitals, each with over five years of clinical experience, ensuring high diagnostic reliability.
- The evaluation setup allows for hierarchical and semantically related diagnoses, accommodating overlapping disease boundaries and reducing the need for strict subtype distinctions—mirroring actual clinical practice.
- A predefined caption schema was used to structure image descriptions, including fields for color, location, shape, lesion type, number, size, texture, border and surface characteristics, distribution, surrounding features, and other relevant descriptors, ensuring consistent and comprehensive annotation.
- During model training, the dataset was split into training and validation sets, with captions processed to align with the schema and filtered to maintain high-quality, clinically relevant descriptions.
- No image cropping was applied; all images were used in their original form to preserve diagnostic details. Metadata was constructed based on the schema, with empty fields left blank when descriptors were not applicable.
Method
The authors leverage a two-stage training pipeline for dermatological diagnosis, structured around an information transmission framework that optimizes the flow of visual information from raw image pixels to final diagnostic outputs. This approach is grounded in the theoretical principle that diagnostic performance is constrained by the efficiency of information recovery within a limited decoding space. The framework decomposes visual information into describable features Id (explicit medical signs) and non-describable features In (implicit pathological textures). Stage I, termed information compression, employs a medical captioning task to force the encoder to prioritize the compression of Id into linguistically interpretable representations, establishing a high-capacity channel for key diagnostic features. Stage II, semantic decoding, then fine-tunes the model to integrate In and decode the joint information into diagnosis-specific semantics, ensuring the final output is grounded in both explicit clinical evidence and implicit visual cues. This staged optimization aims to maximize the recoverable information, approaching the theoretical upper bound of the diagnostic space.
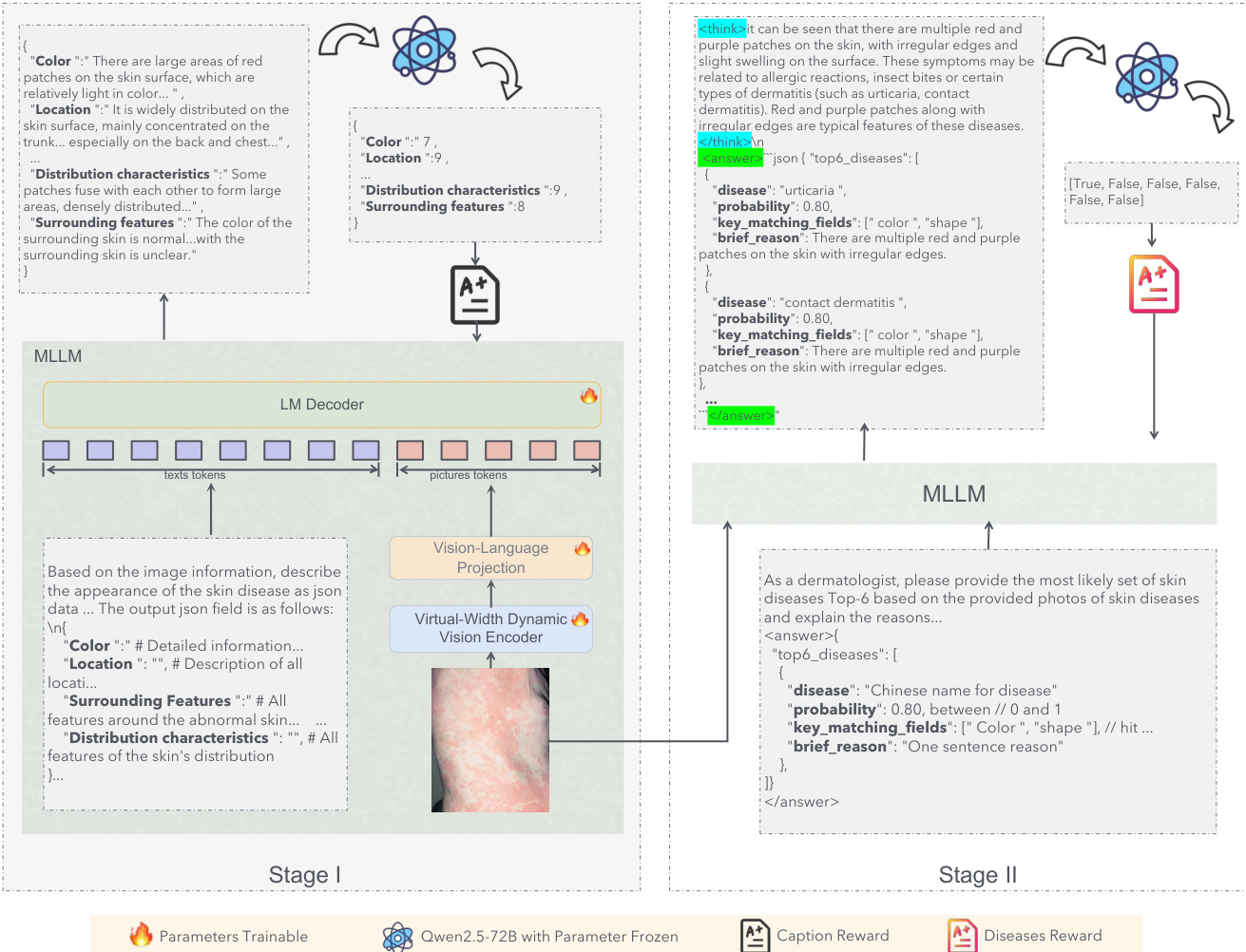
The core of the visual representation is the Dynamic Vision Encoder (DVE), which addresses the capacity limitations of standard Vision Transformers. Standard models rely on static linear layers, y=Wx+b, where the weight matrix W is fixed after training. According to Cover's Theorem, the probability P(N,d) that N random patterns are linearly separable in a d-dimensional space is approximately 1 if N≤2d and 0 if N≫2d. In dermatology, the number of visual patterns N (e.g., texture, erythema, scale) can be very large, while the physical dimension d of the encoder is fixed, leading to a capacity collapse where fine-grained details are averaged out. To circumvent this, the authors propose FDLinear, a mechanism for implicit high-dimensional mapping that achieves virtual dimension expansion without the computational cost of physical expansion. FDLinear decouples the weight space into K orthogonal spectral bases {B1,…,BK}, constructed via frequency disjoint partitioning of the weight matrix's Fourier spectrum. As shown in the figure below, the original weight matrix Wori in the spatial domain is transformed to the Fourier domain via the Discrete Fourier Transform (DFT). The frequency spectrum is partitioned into disjoint groups, such as low-frequency components (e.g., P1) and high-frequency components (e.g., P2). A specific spatial basis Bk is generated by retaining only the learnable parameters in the k-th frequency group and applying an inverse DFT (iDFT), ensuring each basis specializes in a distinct frequency band and minimizes spectral redundancy.
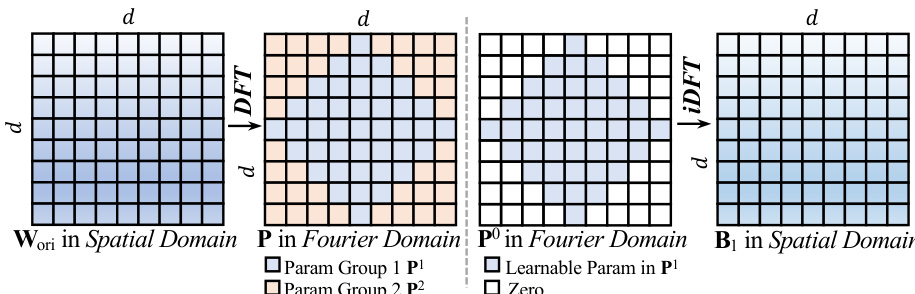
The dynamic weight W(xˉ) is constructed as a context-aware linear combination of these bases, W(xˉ)=∑k=1Kαk(xˉ)⋅Bk, where the dynamic coefficient αk(xˉ) is factorized into three orthogonal modulation vectors predicted by a lightweight fully-connected bottleneck layer. This operation can be viewed as a two-step "Expand-and-Collapse" process. The projection of the input x onto all bases generates a massive hidden representation H∈RK×d, virtually expanding the channel dimension from d to K×d. In this hyperspace, the complex visual manifolds are unfolded and become linearly separable. Crucially, the model never explicitly materializes H; instead, it fuses the aggregation step before the matrix multiplication, computing y=W(xˉ)x, where W(xˉ) is a pre-computed d×d dynamic matrix. This allows the model to enjoy the geometric capacity of a massive width while maintaining the computational footprint of a compact layer.
The training process employs a reinforcement learning (RL) framework, specifically Group Relative Policy Optimization (GRPO), to optimize the model's performance. GRPO operates on groups of candidate outputs, eliminating the need for a separate critic model. For a given query q, a group of G candidate outputs is sampled from the current policy, and their rewards r1,…,rG are computed. Advantages are normalized within the group, Ai=std(rj)ri−mean(rj), to stabilize training. The GRPO objective optimizes a clipped importance-weighted surrogate with a KL penalty to a reference policy, enabling efficient utilization of reward signals from diverse outputs. In Stage I, the model is trained to generate structured medical captions. The reward function is designed to ensure clinical validity and semantic completeness, computing a weighted average of individual attribute scores, R=∑iαi⋅si, where si is the score for attribute i and αi is its weight. The weight of each attribute is determined by its frequency in the LLM's diagnostic reasoning. In Stage II, the model predicts disease categories, and the reward is defined by the positional weight of the correct diagnosis in the ranked list of top-K predictions, encouraging both correctness and high ranking. The model is initialized from Qwen2.5-VL-Instruct-7B, with FDLinear operators replacing static linear layers in the Vision Transformer at specific layers. The first-stage training uses AdamW with a learning rate of 1×10−6, and the second-stage RL training continues from the first-stage checkpoint with a learning rate of 5×10−7, implemented using the VERL framework.
Experiment
-
Visual Verification and Efficiency Analysis: FDLinear validates the "virtual width expansion" hypothesis by successfully unfolding non-linear manifolds (e.g., intertwined spirals, concentric circles) through dynamic, geometry-aware weight directions, enabling compact models to achieve capacity comparable to much larger networks. This results in parameter-efficient storage (<5% overhead) and inference speed on par with standard linear layers, effectively bridging the gap between a 0.6B vision perceiver and a 7B language reasoner.
-
Stage II Dermatological Diagnosis Training: The use of reinforcement learning (RL) for top-K diagnosis training enables robust handling of terminological diversity and efficient learning without exhaustive label enumeration, outperforming supervised fine-tuning in semantic flexibility and scalability.
-
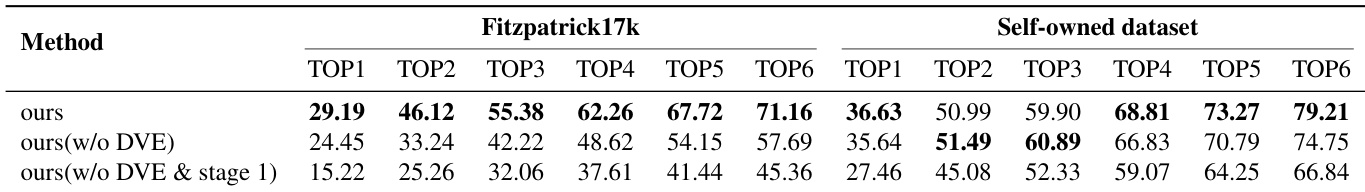
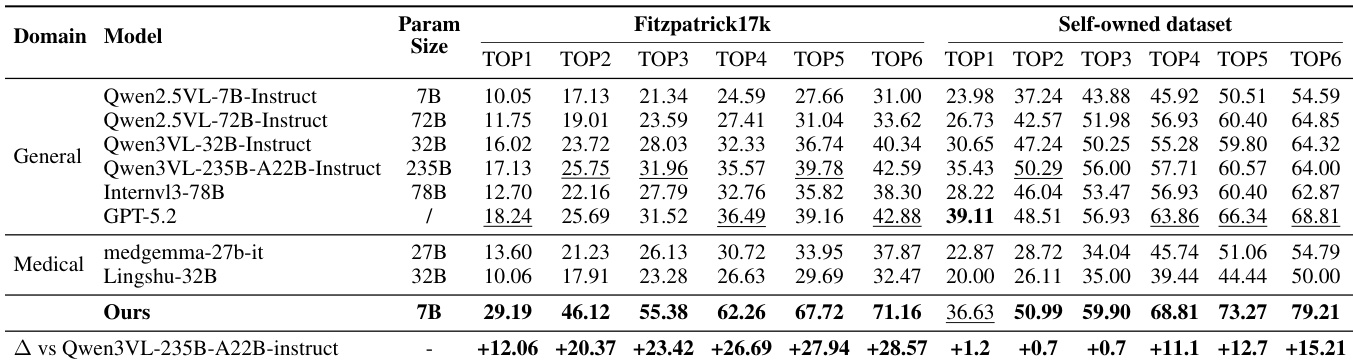
Main Results: On Fitzpatrick17k, the 7B SkinFlow model achieves Top-1 accuracy of 29.19%, surpassing the strongest baseline (GPT-5.2) by 10.95% and Qwen3-VL-235B by 12.06%, with Top-6 accuracy of 71.16%—a +28.57% improvement over Qwen3-VL-235B. On the Self-owned dataset, it reaches 79.21% Top-6 accuracy, significantly outperforming GPT-5.2 (68.81%) and Qwen3-VL-235B (64.00%), demonstrating superior diagnostic candidate pool quality despite being orders of magnitude smaller.
-
Ablation Studies: Stage 1 caption training boosts Top-1 accuracy from 27.46% to 35.64% (Self-owned) and 15.22% to 24.45% (Fitzpatrick17k). Adding the Dynamic Visual Encoding (DVE) module further improves Top-1 accuracy to 36.63% and Top-6 to 79.21%, with a 4.74% gain on Fitzpatrick17k and a 13% increase in Top-6 accuracy, confirming its critical role in fine-grained feature adaptation.
-
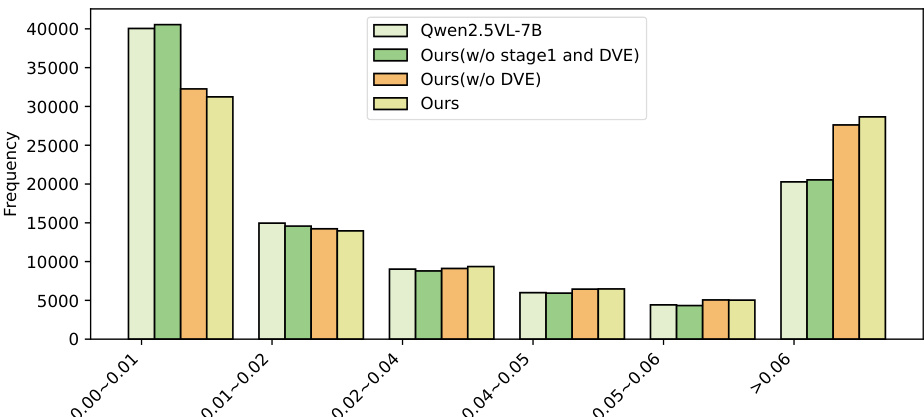
Qualitative Analysis: Visual attention maps show that SkinFlow shifts from diffuse global scanning to precise lesion localization, effectively suppressing background noise. The full model exhibits a high-confidence attention distribution (>0.06), with a 500-sample statistical analysis confirming a significant "shift-to-right" in attention weight distribution, indicating superior signal-to-noise ratio in visual reasoning.
Results show that the full model (Ours) achieves the highest frequency in the >0.06 attention weight bin, indicating the strongest confidence in diagnostic feature selection. The model without Stage 1 training (Ours w/o stage1 and DVE) and the variant without DVE (Ours w/o DVE) exhibit lower frequencies in this high-confidence range, while the baseline Qwen2.5-VL-7B shows a heavy concentration in the 0.00–0.01 bin, reflecting diffuse and uncertain attention. This demonstrates that both Stage 1 caption training and the Dynamic Visual Encoding (DVE) module are essential for suppressing background noise and amplifying diagnostic signal strength.
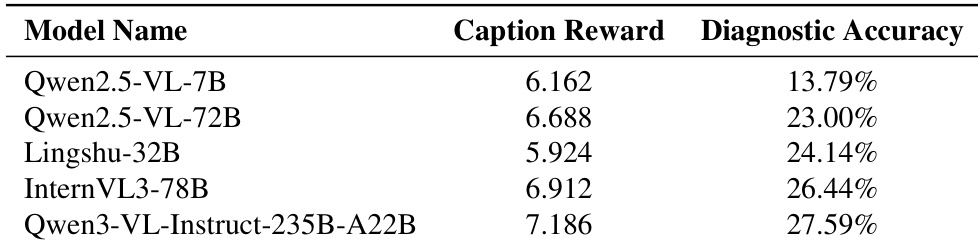
The authors use the table to compare the diagnostic accuracy of several multimodal models, showing that Qwen3-VL-Instruct-235B-A22B achieves the highest accuracy at 27.59%. Results show that models with higher caption rewards generally perform better, with Qwen2.5-VL-72B achieving 23.00% accuracy despite a lower caption reward than some competitors, indicating that caption reward is not the sole predictor of diagnostic performance.
Results show that the proposed SkinFlow model achieves state-of-the-art performance on both Fitzpatrick17k and the Self-owned datasets, outperforming larger general-purpose and medical models despite having significantly fewer parameters. The model's superior accuracy, particularly in Top-6 rankings, demonstrates its effectiveness in generating a robust diagnostic candidate pool, with notable improvements over baselines like GPT-5.2 and Qwen3VL-235B.
Results show that the full SkinFlow model achieves the highest performance across all metrics on both the Fitzpatrick17k and Self-owned datasets, with a Top-1 accuracy of 29.19% and 36.63% respectively. The ablation study demonstrates that both the dynamic visual encoding (DVE) module and the Stage 1 captioning task are critical, as removing either component significantly reduces performance, particularly on the more challenging Fitzpatrick17k dataset.