Command Palette
Search for a command to run...
Meta AI Et al. Proposed a New Protein Dynamic Fusion Characterization Framework, FusionProt, Which Enables Iterative Information Exchange and Achieves SOTA Performance in Multiple tasks.
Proteins are the executors of life functions, and their mysteries lie in two dimensions:One is the one-dimensional (1D) sequence formed by amino acids connected end to end, and the other is the three-dimensional (3D) structure formed by the folding and winding of the sequence.Previous models typically specialize in either one, either mastering "sequence language" like protein language models (PLMs) like ProteinBERT and ESM, or discerning "structural morphology" like three-dimensional protein representation technologies like GearNet. Even when models attempt to combine the two, they often employ a simplified, spliced approach, seemingly allowing each specialist to operate independently rather than collaboratively.
In this context, the research teams of Technion-Israel Institute of Technology and Meta AI jointly proposed the protein representation learning framework FusionProt.It aims to simultaneously learn a unified representation of protein one-dimensional sequence and three-dimensional structure.This research innovatively introduces a learnable fusion token as an adaptive bridge between the protein language model (PLM) and the 3D structure graph, enabling iterative information exchange between the two. FusionProt has achieved state-of-the-art performance in a variety of protein-related biological tasks.
The related research was published on bioRxiv under the title "FusionProt: Fusing Sequence and Structural Information for Unified Protein Representation Learning."
Research highlights:
* The FusionProt framework breaks through the limitations of previous structure fragmentation processing by effectively integrating one-dimensional and three-dimensional modalities, and improves the accuracy of capturing protein functions and interaction properties.
* A novel cross-modal fusion architecture that uses learnable fusion tokens to enable iterative information exchange between the protein language model (PLM) and protein 3D structure graphs.
* FusionProt achieves state-of-the-art performance in multiple protein tasks and demonstrates the model's potential for application in real-world biological scenarios through case studies.
Paper address:
Follow the official account and reply "FusionProt" to get the complete PDF
More AI frontier papers:
https://hyper.ai/papers
Systematically construct datasets using public protein databases
In this study, the research team made full use of public protein databases and ensured the effectiveness of FusionProt in multiple protein understanding tasks through systematic dataset construction, strict data partitioning strategy and multi-task evaluation framework.
In the pre-training stage, the study used the protein structure database (AlphaFold DB) as the core data source.The database contains 805,000 high-quality three-dimensional protein structures predicted by AlphaFold2. The researchers selected this dataset primarily for the following reasons: Firstly, AlphaFold2 is currently recognized as a state-of-the-art model in protein structure prediction, and its predictions are highly reliable, effectively reducing reliance on the quality and availability of external experimental structural data; secondly, using a unified predicted structure ensures consistency across data sources, facilitating fair comparison with previous state-of-the-art work such as SaProt and ESM-GearNet.
The research team further systematically evaluated the model's performance across three authoritative downstream tasks. The tasks used datasets from DeepFRI, which provides authoritative data partitioning and uses Fmax as a unified evaluation metric, comprehensively measuring the model's performance in enzyme function annotation and gene ontology inference. The Mutational Stability Prediction (MSP) task used the same dataset and evaluation protocol as ESM-GearNet, using AUROC as the evaluation metric to assess the model's ability to predict the effects of mutations on protein complex stability.
Iterative information exchange mechanism driven by "fusion token"
The design of FusionProt revolves around a core idea:Through learnable fusion tokens, it acts as a bridge for bidirectional, iterative cross-modal interaction between protein sequence and structure.This achieves deep fusion and unified representation of the two types of information.
First, the framework is built on the basis of the "sequence-structure" dual-modal parallel encoding architecture. At the sequence level, ESM-2 is used as the protein language model to encode the amino acid sequence; at the structural level, the GearNet encoder is used as the structural model to model the protein's three-dimensional structure diagram. The learnable fusion token dynamically shuttles between the two modalities during the training process, enabling iterative exchange and fusion of information. At the sequence level, it is connected to the protein sequence, and the amino acid will query the relevant unique fusion token to extract and integrate valuable information. At the structural level, it is added to the protein three-dimensional graph and merged and connected as a node. The structural layer is processed by a message passing neural network, enabling the fusion token to integrate global spatial structural information.
Secondly, the core driving force of this framework lies in the iterative fusion algorithm.The process involves splicing the fusion tokens into the updated sequence, then passing them to the structure layer and feeding them into the structure graph network as new nodes. The updated fusion tokens are then fed back to the sequence layer for the next round of interaction. This iterative process aligns and adjusts the different modal spaces through learnable linear transformations. Through this iterative process, the model representations are combined to form a unified and rich protein representation.
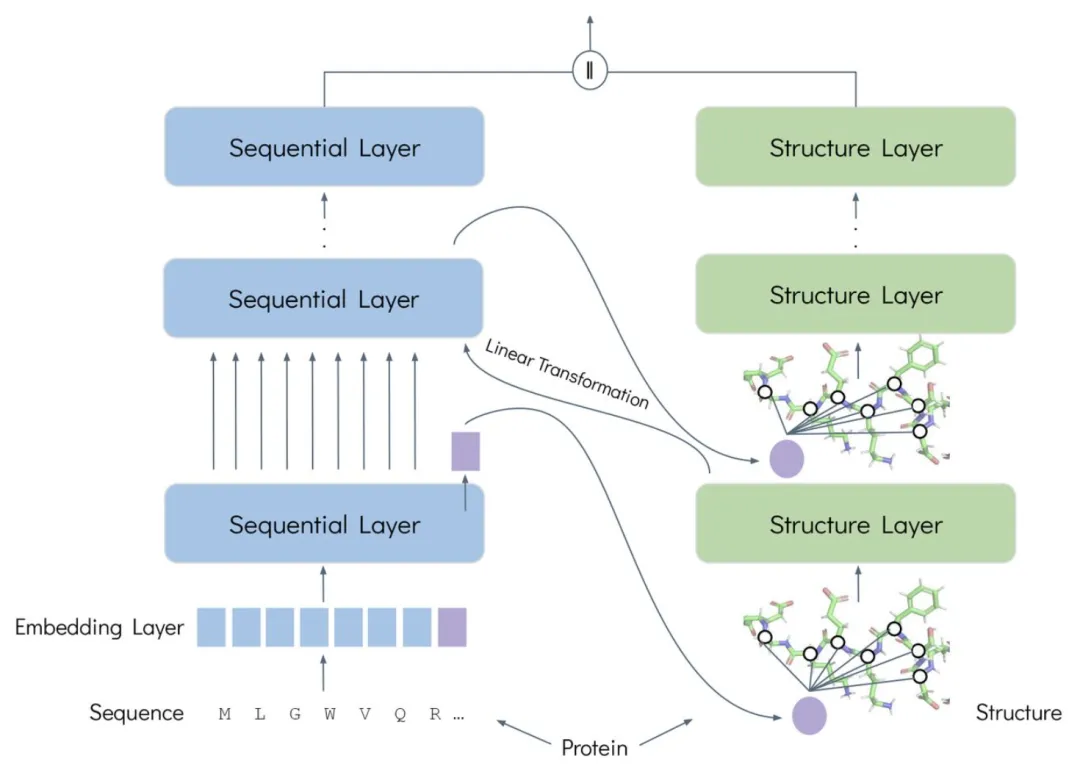
Finally, FusionProt uses Multiview Contrastive learning as a pre-training objective.A diverse view is constructed by randomly selecting consecutive subsequences and hiding the graph edges of 15%. The InfoNCE loss function is then used to align representations in the latent space, maintaining the similarity of related protein subcomponents when mapped to the low-dimensional latent space. In implementation, the research team performed pre-training on the AlphaFold DB database mentioned above. During pre-training, FusionProt used a learning rate of 2e-4, a global batch size of 256 proteins, and conducted 50 rounds of training. The input sequence was truncated to 1,024 tokens to accommodate long protein sequences. Furthermore, fine-tuning was performed by adding task-specific classification head predictions and evaluating the same hyperparameters as the latest SaProt model. All experiments were performed on 4 x NVIDIA A100 80GB GPUs, with a single pre-training session taking approximately 48 hours.
Comprehensively surpassing existing SOTA, the fusion mechanism has significant effects
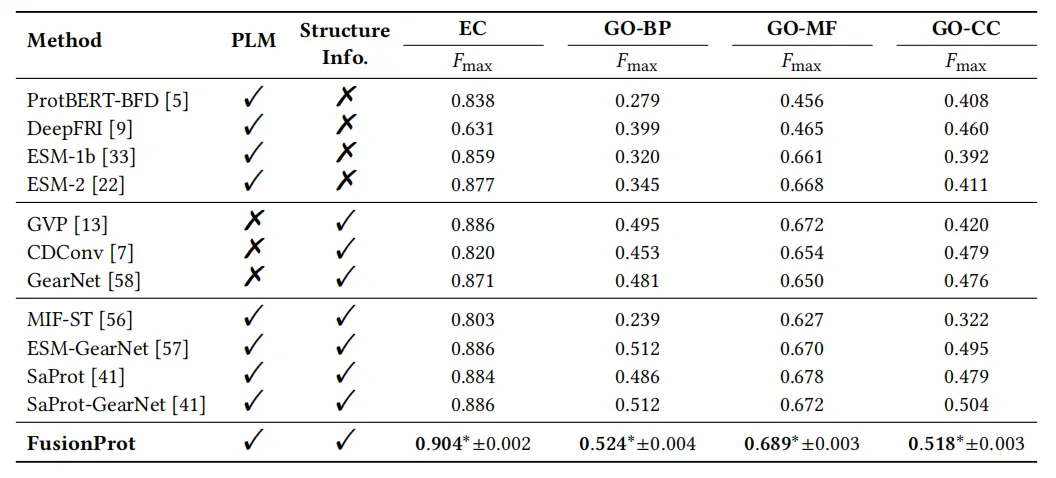
The study was extensively tested in multiple downstream tasks.The results show that the FusionProt framework achieves SOTA performance in multiple benchmarks.The experimental results are shown in the figure below.
In the EC number prediction evaluation, the research team compared the performance of FusionProt with 11 baseline models. The results showed that FusionProt achieved the highest Fmax = 0.904, significantly outperforming models that rely solely on sequence (such as ProtBERT-BFD, 0.838, ESM-2, 0.877), and also surpassing GearNet (0.871) that only uses structural information. At the same time, compared with other methods that attempt to utilize these two types of information (such as MIF-ST, ESM-GearNet, etc.), it still takes the lead. This result shows that compared to simply using one modality as the context of another modality, FusionProt's iterative fusion mechanism can more fully retain key three-dimensional structural information, thereby more accurately capturing the subtle structural differences that catalytic activity depends on.
In the GO term prediction evaluation, FusionProt achieved the best results in the three subtasks of biological process, molecular function, and cellular component, once again demonstrating the effectiveness of learnable fusion tokens in joint modeling of sequence and structure.
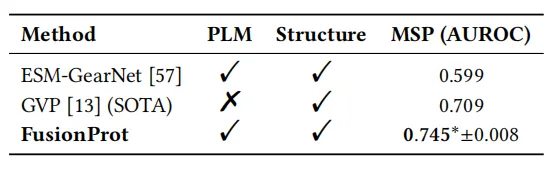
The research team also conducted an evaluation of mutational stability prediction. The experimental results showed that FusionProt achieved the highest AUROC of all evaluated methods, with statistical significance (p < 0.05). This performance significantly improved by 5.11 TP3T compared to the current state-of-the-art method, GVP, highlighting the effectiveness of its iterative fusion mechanism in integrating long-range sequence-structure dependencies. Furthermore, FusionProt enables bidirectional cross-modal interaction through learnable fusion tokens, making protein representation more expressive and biologically grounded.
To evaluate the effectiveness of FusionProt's key designs, the research team conducted further ablation experiments. The team tested the system at different fusion injection frequencies and found that optimal performance was achieved when the fusion markers performed multiple rounds of interactions between the sequence and structure encoders at a standard frequency; while reducing the interaction frequency significantly weakened the performance.This shows that frequent information exchange is crucial for capturing cross-modal dependencies.
Finally, in a biological case analysis, FusionProt successfully predicted the EC number of the RNA polymerase ω subunit protein, which is difficult to handle with traditional methods. This result completely failed in models such as ESM-2, further proving that the learned representation can capture complex "structure-function" relationships and show broad application potential in drug development and protein function analysis.
Cross-modal fusion has become an obvious trend
FusionProt has paved a new path for protein representation learning, demonstrating that the "language" and "morphology" of proteins should communicate with each other, not with each other. With the continuous advancement of artificial intelligence in life sciences, cross-modal fusion has become a clear trend.
Westlake University proposed the concept of a structure-aware vocabulary and combined amino acid residue tokens with structure tokens to train a large-scale universal protein language model, SaProt, on a dataset of approximately 40 million protein sequences and structures. This model comprehensively outperformed established baseline models across 10 important downstream tasks. The related research, titled "SaProt: Protein Language Modeling with Structure-aware Vocabulary," was selected for ICLR 2024.
Paper address:
https://openreview.net/forum?id=6MRm3G4NiU
A joint study titled "Structure-Aligned Protein Language Model" published by the University of Montreal and Mila proposes a structure-aligned protein language model that incorporates structural information using contrastive learning. By optimizing the model's predicted structural tokens, the model significantly improves the performance of protein contact prediction tasks.
Paper address:
https://arxiv.org/abs/2505.16896
Get high-quality papers and in-depth interpretation articles in the field of AI4S from 2023 to 2024 with one click⬇️