Command Palette
Search for a command to run...
ACL 2025: Oxford University and Others Propose Medical GraphRAG, Setting a New Record for question-answering Accuracy and Achieving SOTA Results on 11 Datasets

The medical field's knowledge system is built on thousands of years of discovery and accumulation, covering a vast amount of principles, concepts, and practical norms. Effectively adapting this knowledge to the limited context window of current large language models presents insurmountable technical obstacles. Although supervised fine-tuning (SFT) offers an alternative, due to the closed-source nature of most commercial models, this approach is not only costly but also extremely impractical in practice. In addition, the medical field places extremely high demands on the accuracy of terminology and the rigor of facts. For non-professional users, verifying the accuracy of large models for medical-related answers is itself an extremely challenging task. Therefore, how to enable large models to utilize large external datasets for complex reasoning in medical applications and generate accurate and credible answers supported by verifiable sources has become a core issue in current research in this field.
The emergence of retrieval enhanced generation (RAG) technology provides a new approach to solving the above problems.It can respond to user queries using specific or private datasets without further training of the model.However, traditional RAGs still fall short when it comes to synthesizing new insights and handling tasks that require a holistic understanding of a broad range of documents. The recently proposed GraphRAG significantly outperforms classic RAGs in complex reasoning by leveraging LLMs to construct a knowledge graph from raw documents and then retrieving knowledge based on the graph to enhance answers. However, GraphRAG's graph construction lacks specific design to ensure answer authentication and credibility, and its hierarchical community construction process is costly due to its general purpose nature, making it difficult to directly and effectively apply to the medical field.
To address this situation, a joint team from the University of Oxford, Carnegie Mellon University, and the University of Edinburgh proposed a graph-based RAG method specifically for the medical field - Medical GraphRAG (MedGraphRAG).This method effectively improves the performance of LLM in the medical field by generating evidence-based answers and official medical terminology explanations, which not only enhances the credibility of the answers but also significantly improves the overall quality.
The related research results, titled "Medical Graph RAG: Towards Safe Medical Large Language Model via Graph Retrieval-Augmented Generation", have been selected for ACL 2025.
Research highlights:
* This study proposed for the first time the Tukey RAG framework specifically for use in the medical field.
* This study developed a unique triple graph construction and U-retrieval method to enable the Large Language Model (LLM) to efficiently utilize the entire RAG data and generate evidence-based answers.
* MedGraphRAG outperforms other retrieval methods and fine-tuned medical-specific large language models on multiple medical question answering benchmarks.
Paper address:
More AI frontier papers:
Research based on three types of data
The data used in this study are divided into three categories, with characteristics of each type appropriate to its role in the study:
* RAG data
Considering that users may use frequently updated private data (such as patient electronic medical records), the study selected the public electronic health record dataset MIMIC-IV, which can simulate the dynamically changing private data scenarios in actual applications and provide a basis for verifying the practicality of the method.
* Repository data
This dataset is used to provide trusted sources and authoritative vocabulary definitions for the answers of the large model. The upper-level repository data is MedC-K, which contains 4.8 million biomedical academic papers, 30,000 textbooks, and all evidence publications from FakeHealth and PubHealth. Its content covers a wide range and is academically authoritative; the underlying repository data is the UMLS graph, which contains authoritative medical vocabulary and semantic relationships to ensure the accuracy of medical terminology.
* Test data
This dataset is used to evaluate the performance of the MedGraphRAG method, including the test part of 9 multiple-choice biomedical datasets in MultiMedQA (including MedQA, MedMCQA, PubMedQA, MMLU clinical topics, etc.), which is used to test the performance of the method in routine medical question answering; 2 public health fact verification datasets FakeHealth and PubHealth are used to evaluate the evidence-based answering ability of the method; in addition, the study also collected the DiverseHealth test set, which contains 50 real clinical questions covering a wide range of topics such as rare diseases and minority health, focusing on health equity, which can further enrich the evaluation dimensions.
MedGraphRAG: Based on sliding window partitioning, label clustering and U-search
As shown in the figure below,The overall workflow of MedGraphRAG mainly includes three core links:Build a knowledge graph based on documents, organize and summarize the graph to support retrieval, and respond to user queries by retrieving data.
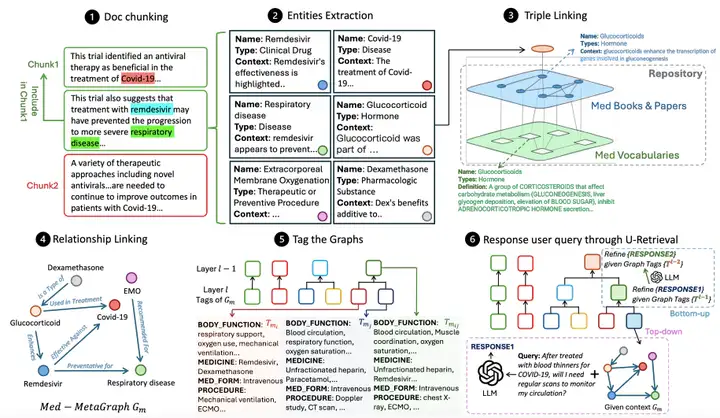
Medical Graph Construction first performs semantic document chunking, dividing the document into data chunks that conform to the LLM context constraints.The study adopts a hybrid method that combines character separation and topic semantic segmentation, that is, first separating paragraphs by line breaks, and then constructing LLM LG through the graph to judge the topic relevance between the paragraph and the current block to decide whether to split the block.At the same time, a 5-segment sliding window is introduced to reduce noise, and the LG tag restriction is used as the hard threshold for block segmentation, taking into account both semantic logic and model context constraints.
After block division, it enters the entity extraction process. With the help of LG with entity extraction prompts, relevant entities are identified from each block, and a structured output containing name, type and context description is generated, paving the way for subsequent entity linking.Triple Linking is the key to ensuring accuracy.A repository graph (RepoGraph) is constructed to link user RAG documents with trusted sources: the bottom layer is the UMLS graph (Med Vocabularies) containing medical vocabulary and relationships, and the upper layer is constructed from medical textbooks and scholarly articles (Med Books & Papers). Next, the researchers define entities extracted from the RAG documents as E1. Based on the inter-entity correlation, these entities are linked to entities E2 extracted from medical books or papers. E2 is further linked to UMLS entities E3, forming a triple structure of [RAG entity, source, definition], ensuring that each entity can be traced back to a clear source and standard definition. Relationship linking is then performed. An LG with relationship recognition hints identifies relationships between RAG entities based on entity content and references, generating phrases containing source entities, target entities, and relationship descriptions. Finally, a directed meta-medical graph is generated for each data block.
After the graph is built, you need to tag the graphs to improve retrieval efficiency.Unlike GraphRAG's costly graph community construction approach, this method leverages the structured nature of medical text and summarizes each meta-medical graph using predefined labels (such as symptoms, medical history, physical function, and medications) to generate a structured label summary. This method then uses dynamic threshold agglomerative hierarchical clustering based on label similarity to group the graphs and generate a more abstract, comprehensive label summary. Initially, each graph is treated as an independent group. Label similarity between cluster pairs is iteratively calculated, and the top 20% cluster pairs with the highest similarity are merged to generate a new label summary layer. This process is limited to 12 layers, achieving a balance between accuracy and efficiency.
The final U-retrieval stage achieves efficient query response by responding to LLM LR.First, LR generates a label summary for the user query. Through top-down precision retrieval, starting from the top-level label, it matches the most similar labels layer by layer, locating the target meta-medical graph. Based on the embedding similarity between the query and the entity content, it retrieves the top-ranked entities and their nearest triplet neighbors, and uses these entities and relationships to generate an initial answer. Next, the bottom-up answer refinement phase begins. LR adjusts the answer based on the previous layer's label summary. This process repeats until the target level (typically 4-6 layers) is reached, ultimately generating an answer that balances global context awareness and retrieval efficiency.
MedGraphRAG: Validated on 6 models and 11 datasets to achieve SOTA
To verify the performance of MedGraphRAG, the study selected six large language models and designed multiple sets of experiments, including Llama2 (13B, 70B), Llama3 (8B, 70B), Gemini-pro and GPT-4.The main comparison is with the standard RAG implemented by LangChain and GraphRAG implemented by Microsoft Azure.All methods are run on the same RAG data and test data.
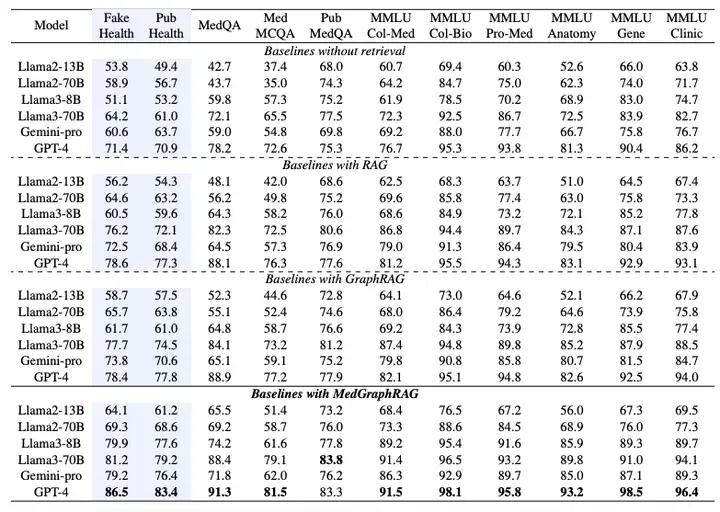
As shown in the table below, the performance of the Multi-Choice Evaluation is measured by the accuracy of selecting the correct option.Experimental results show that MedGraphRAG significantly outperforms the baseline without retrieval function, standard RAG, and GraphRAG:Compared to a no-search baseline, it achieved an average improvement of nearly 101 TP3T in fact-checking and 81 TP3T in medical question answering. Compared to GraphRAG, it achieved an improvement of approximately 81 TP3T in fact-checking and 51 TP3T in medical question answering. The improvement was even more significant for smaller models (such as Llama2 13B), demonstrating its effective integration of model reasoning capabilities and external knowledge. When applied to larger models (such as Llama70B and GPT-4), it achieved state-of-the-art performance on 11 datasets, even surpassing models fine-tuned on medical corpora such as Med-PaLM 2 and Med-Gemini, establishing a new state-of-the-art performance on the medical LLM leaderboard.
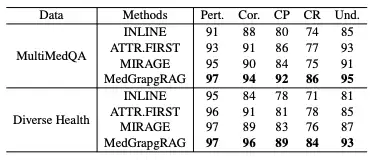
In the Long-form Generation Evaluation,This study compared MedGraphRAG with models such as Inline Search and ATTR-FIRST in terms of five dimensions: relevance, correctness, citation precision, citation recall, and comprehensibility on the MultiMedQA and DiverseHealth benchmarks.The results are shown in the table below. MedGraphRAG scored higher in all indicators, especially in citation precision, recall, and comprehensibility, thanks to its evidence-based answers and clear explanations of medical terms.
In the case study, faced with a complex case of chronic obstructive pulmonary disease (COPD) and heart failure, GraphRAG's recommendations ignored the impact of drugs on heart failure, while MedGraphRAG was able to recommend safe drugs. This was due to the direct link between its entities and references, avoiding the omission of key information caused by the interweaving of information in GraphRAG.
Integration of Knowledge Graph and Large Language Model
In the intersection of medicine and artificial intelligence, the integration of knowledge graphs and large language models is becoming a key direction for promoting technological breakthroughs, providing new ideas for solving complex problems in the medical field.
For example, the KG4Diagnosis framework proposed by a joint team from Cambridge University and Oxford University,It simulates real-world medical systems through a hierarchical multi-agent architecture and combines knowledge graphs to enhance diagnostic reasoning capabilities, covering automated diagnosis and treatment planning for 362 common diseases.A research team from Fudan University has comprehensively mapped the proteome of human health and disease. By deeply analyzing the plasma proteome data of 53,026 individuals over a median follow-up period of 14.8 years, the map covers 2,920 plasma proteins and 406 pre-existing diseases, 660 newly-emerged diseases during follow-up, and 986 health-related phenotypes.Uncovering numerous protein-disease and protein-phenotype associations,Provide important basis for precision medicine and new drug development.
The AMIE system launched by Google DeepMind,Integrate the long-context reasoning capabilities of the Gemini large model with the knowledge graph,By dynamically searching clinical guidelines and drug knowledge bases, a consistent management plan can be generated across multiple diagnosis cases. For example, for patients with chronic obstructive pulmonary disease (COPD) and heart failure, cardioselective beta-blockers can be accurately recommended, avoiding the drug interaction risks of traditional AI systems.
The biomedical knowledge graph built by AstraZeneca integrates 3 million documents and internal research data, and accelerates the screening of new drug candidates by analyzing the drug-target-disease association network.The map not only includes the indications of approved drugs, but also covers the "off-label use" data in clinical trials.Providing decision support for the repurposing of established drugs. Furthermore, IBM Watson Health's knowledge graph platform integrates 1 billion pieces of patient data with evidence-based guidelines to generate personalized lung cancer treatment plans that include genetic testing, drug sensitivity prediction, and follow-up plans, reducing the predicted error in patient survival to ±2.3 months.
These innovative practices not only drive the iterative upgrade of medical AI technology but also demonstrate tremendous potential in improving diagnostic accuracy, accelerating drug development, and optimizing clinical decision-making. As the technology continues to mature, the integration of knowledge graphs and large language models will further break down information barriers in the medical field and inject sustained momentum into the development of global healthcare.
Reference articles:
1.https://mp.weixin.qq.com/s/WhVbnoso2Jf2PyZQwV93Rw
2.https://mp.weixin.qq.com/s/RWy4taiJCu3kMPfTzOWYSQ
3.https://mp.weixin.qq.com/s/lMLk








