Command Palette
Search for a command to run...
Model Parameters Exceed RFdiffusion by 5 Times! NVIDIA and Others Release Proteina, Which Achieves SOTA Performance in De Novo Protein Backbone Design

Since the last century, scientists have been exploring the prediction of protein structure based on amino acid sequences, with the vision of using amino acids to create new proteins and build the blueprint of life. However, this grand mission has progressed slowly over the long course of time. It was not until recent years, with the rapid development of AI technology, that it was injected with strong momentum and entered the fast lane of development.
Since 2016, a technological revolution initiated by Xu Jinbo, founder and chief scientist of Molecular Heart, and others has been quietly changing this field. They pioneered the introduction of the deep residual network ResNet architecture into the field of structure prediction.Significant improvement in protein residue contact prediction was successfully achieved.This breakthrough has laid a solid foundation for the deep integration of AI and protein design. After that, many scientific research teams have continued to work hard in this field. A large number of algorithms combining co-evolution and deep learning have emerged. Among them, a series of blockbuster results by David Baker, the 2024 Nobel Prize winner in Chemistry, and AlphaFold have become famous, pushing research in this field to new heights.
However, reviewing previous studies,Unconditional protein structure generation models are often only trained on small-scale datasets, with the number of structures not exceeding 500,000.Moreover, during the synthesis process, the neural networks of these models lack effective control methods, and there is a large gap in both scale and performance compared with generative models in fields such as natural language, image or video generation.
In the fields of natural language, image, and video generation, people have witnessed earth-shaking changes and major breakthroughs due to scalable neural network architectures, large-scale training data sets, and sophisticated semantic control. This has made researchers ponder: Can we learn from the successful experience in these fields and make similar extensions and controls for protein structure diffusion and flow models, so as to achieve a qualitative leap in the field of protein design?
What is gratifying is that NVIDIA recently collaborated with Mila, the Quebec Artificial Intelligence Research Institute, the University of Montreal, and the Massachusetts Institute of Technology to develop a new type of large-scale flow protein backbone generator, Proteina. Proteina has five times the number of parameters of the RFdiffusion model and expands the training data to 21 million synthetic protein structures.We achieve state-of-the-art performance in de novo protein backbone design and generate diverse and designable proteins of unprecedented length—up to 800 residues.
The related research results, titled "Proteina: Scaling Flow-based Protein Structure Generative Models", have been selected for ICLR 2025 Oral.

Paper address:
https://openreview.net/forum?id=TVQLu34bdw&nesting=2&sort=date-desc
Recommend an academic sharing event. The latest Meet AI4S live broadcast invitation is at 12:00 noon on March 7.Huang Hong, associate professor at Huazhong University of Science and Technology, Zhou Dongzhan, young researcher at AI for Science Center of Shanghai Artificial Intelligence Laboratory, and Zhou Bingxin, assistant researcher at the Institute of Natural Sciences of Shanghai Jiao Tong University,Introduce personal achievements and share scientific research experience.
AI empowers protein design: from structure to sequence, from prediction to design
In the process of life science research, protein design has always occupied an extremely critical position. For a long time, learning rules and patterns from massive amounts of protein sequence data has been a pain point faced by researchers. Fortunately, with the support of AI technology, this field has taken the lead in ushering in a turning point.
For example,AlphaFold3 launched by DeepMind improves the modeling of DNA, RNA and small molecule interactions.The ability to accurately predict the structure of protein complexes provides strong support for understanding the complex interactions of proteins within cells. Meta once launched ESMFold, which combines language model with structure prediction.This greatly improves the prediction speed, allowing researchers to obtain protein structure information more efficiently.Microsoft's latest BioEMU-1 simulates the dynamic changes of protein conformation.It has opened up a new avenue for in-depth exploration of the movement mechanism of proteins and drug design.
With these foundations, AI has gradually begun to penetrate into protein structure design.
Protein structure design is mainly based on known protein structures, which are transformed and optimized through various methods to obtain proteins with specific functions or properties. Since the function of a protein is mainly determined by its three-dimensional conformation,The method of directly modeling structural distribution has gradually become the mainstream trend, among which algorithms based on diffusion models or flow models are particularly outstanding.For example, the Chroma model developed by Generate Bio is the first large-scale application of diffusion modeling for precise protein design.Capable of producing "proteins that do not exist at all in nature."
also,RFdiffusion proposed by David Baker can generate protein skeletons with specific functions by fine-tuning the RoseTTAFold structure prediction network.It provides a precise structural basis for the design of functional proteins. Genie2 proposed by researchers from Columbia University and Rutgers University extends the training data to AFDB.Capable of generating complex proteins with multiple independent functional sites.
As we all know, the structure and sequence of proteins are interrelated. The structure determines the function, and the sequence is the basis of the structure. When the protein structure is changed by AI technology, the protein sequence will inevitably change as well. Protein sequence design is mainly based on the known protein structure, through calculation and prediction methods, to design an amino acid sequence that matches the structure.
At present, AI protein sequence design is mainly divided into two types:One is a fixed backbone protein sequence design tool,For example, the ESM-IF launched by Stanford University uses a paradigm that combines pre-training and fine-tuning, cleverly integrating structural knowledge into functional protein design, providing a strong guarantee for designing proteins with specific functions. The ProteinMPNN proposed by David Baker is based on a graph neural network and can generate matching amino acid sequences based on the main chain structure, providing an efficient and accurate method for protein sequence design.
The other is a function-oriented protein sequence design tool.For example, ProGen, launched by Salesforce, is a conditional generation model that can customize protein sequences according to specific functional requirements, providing a highly flexible solution for the design of functional proteins. ZymCTRL, launched by the University of Girona in Spain, achieves function-oriented design by fine-tuning the pre-trained language model, providing strong support for the precise regulation of protein functions. P450Diffusion proposed by the Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, generates P450 enzyme variants with specific catalytic functions based on the diffusion model, bringing new development opportunities to the field of enzyme engineering.
However, compared with the other three types of protein models, the scale of current protein structure design models is generally small. Specifically, the scale of AlphaFold 3's training set is close to 100 million, BioEmu-1 uses more than 200 million protein sequences from the AFDB database in the pre-training stage, and the number of parameters of ProGen is as high as 1.2 billion. However, RFdiffusion, an outstanding representative in the field of protein structure design, only has its training data from tens of thousands of real protein structures in the Protein Data Bank (PDB) repository, and the total length of the structure that can be generated can only reach 600 amino acid residues. The largest data set of Genie2 is only about 600,000 synthetic structure proteins.
In this context,The industry is eagerly looking forward to the birth of a protein structure design model with larger training data volume, longer total structure length and stronger controllability - Proteina.
Proteina model: a new breakthrough in protein design using AI technology
Proteina, as a flow-based protein structure model, uses an innovative scalable non-equivariant Transformer architecture, which is inspired by the diffusion Transformer in the visual field. It can achieve top performance even without relying on the computationally expensive triangle layer.This enables Proteina to be trained on up to 21 million protein structures, a 35-fold increase in training data, ultimately generating backbones of up to 800 residues.While maintaining designability and diversity, it is significantly better than all previous work.
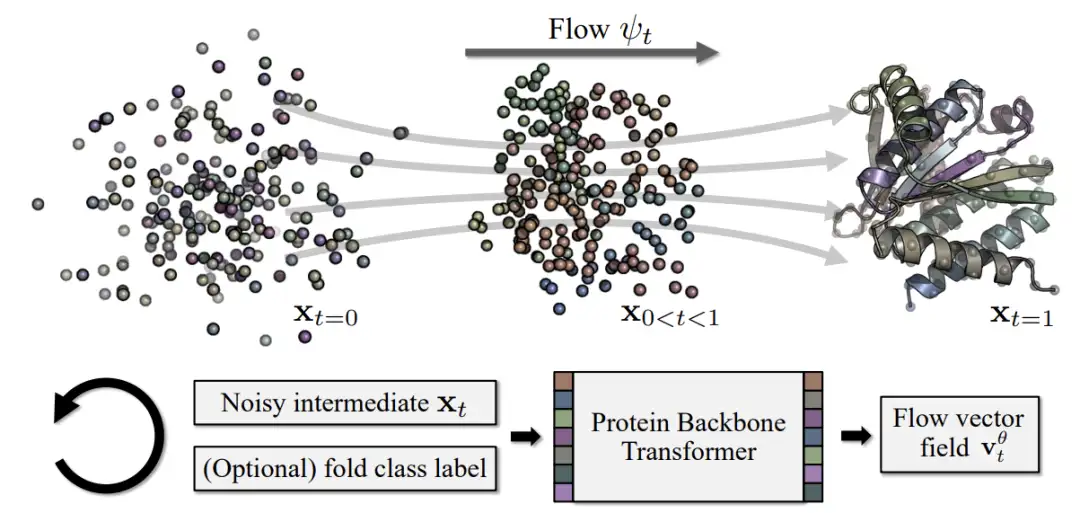
As shown in the figure below, this study mainly uses the Foldseek AFDB clustering DFS dataset used by Genie2.The dataset covers approximately 600,000 synthetic structural proteins.At the same time, the study also used the high-quality filtered AFDB subset D21M filtered from about 214 million AFDB structures.This subset contains approximately 21 million synthetic structural proteins.
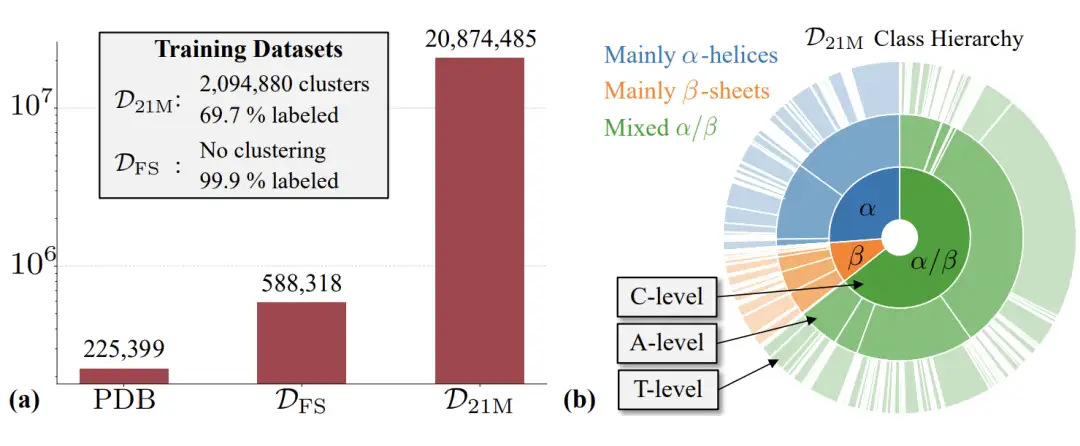
Based on the above two data sets, the researchers further trained three Proteina models: the first is the MFS model, which contains a Transformer with 200 million parameters and a triangular layer with 10 million parameters; the second is the Mno-triFS model, which only contains a Transformer with 200 million parameters, but does not contain any triangular layer or pairwise representation updates; the third is the M21M model, which contains a Transformer with 400 million parameters and a triangular layer with 15 million parameters.
In the field of unconditional protein structure generation, equivariant methods have long dominated, but Proteina has demonstrated that large-scale non-equivariant flow models can also be successful. The trained version has more than 400 million parameters.More than 5 times larger than RFdiffusion, it is currently the largest protein backbone generator.The results also show that models trained on DFS exhibit higher diversity, but researchers can also create much larger amounts of high-quality data from fully synthetic structures than DFS.
In terms of evaluation indicators, Proteina is not satisfied with the traditional diversity, novelty and designability evaluation, but introduces innovative evaluation indicators - directly inputting the empirical labels of DFS into the model. This move enforces diversity between different folding structures and provides unprecedented control over synthetic protein structures through novel folding category condition constraints.
As shown in the figure below, compared with unconditional generation, Proteina's conditional model achieves the most advanced TM-Score diversity while achieving the best FPSD, fS, and fJSD scores.This fully demonstrates its advantage in folding structure diversity "fS" and the better distribution match between the generated structure and the reference data.
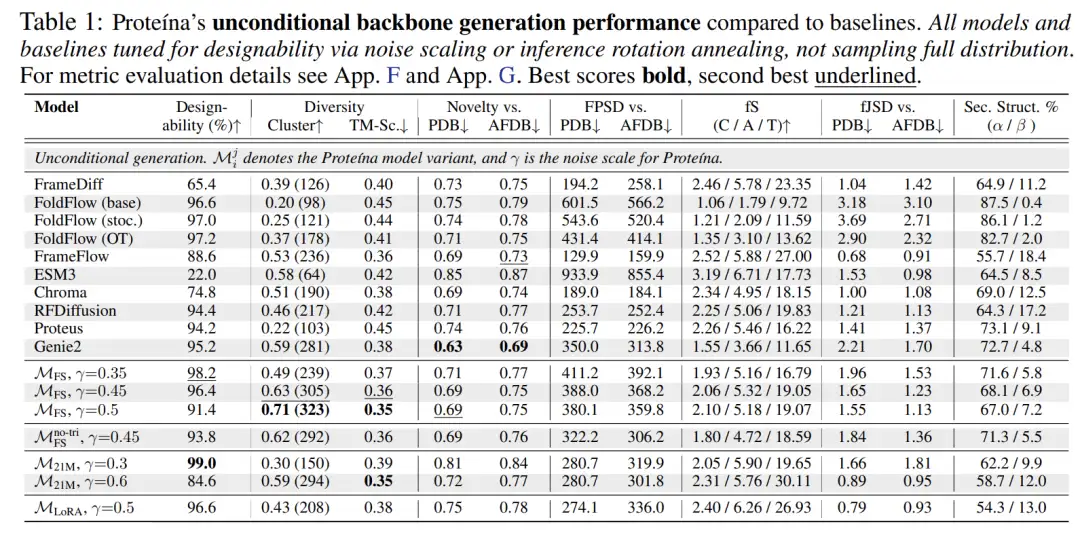
In addition, Proteina adapted the flow matching objective to accommodate protein structure generation and explored phased training strategies, such as using LoRA to fine-tune the model to enable it to generate natural, designable proteins. It also developed new guidance schemes for hierarchical folding category conditional constraints and successfully demonstrated self-guidance to enhance the designability of proteins.In terms of protein backbone generation performance, Proteina has reached SOTA level, especially in long chain synthesis.It significantly outperforms all baseline models and demonstrates superior control capabilities over previous models through novel folding category condition constraints.
Innovations emerging in China’s AI protein design field
At present, with DeepSeek once again detonating the big language model, the field of protein design will undoubtedly usher in new development opportunities, and more and more Chinese forces will emerge. In fact, up to now, in terms of protein structure design alone, Chinese researchers and enterprises have already produced many achievements.
In 2022,Shanghai Tianrang XLab, driven by AI, has launched a new protein design platform - TRDesign. TRDesign can accurately explore all potential possibilities in the protein folding space by learning a lot about the relationship between protein sequence and structure, reversely map the sequence-structure-function association learned from protein folding, and perform end-to-end protein design, testing, stability, and affinity optimization from scratch, thereby designing protein structures that better meet demand.
In 2023,Professor Jinbo Xu, founder of Molecular Heart, launched the NewOrigin big model at the 2023 World Artificial Intelligence Conference "WAIC".By learning from trillions of multimodal big data, the model can achieve multimodal directed generation. A single model can meet the full process requirements of protein generation, including sequence generation, structure prediction, function prediction, and de novo design, solve the problem of generating specific functional proteins required for industrial applications, and evaluate the effect and value in a real industrial environment.
In April 2024, Wuxi Tushen Zhihe Artificial Intelligence Technology Co., Ltd. joined hands with several research institutions toJointly released China's first natural language text-protein large model TourSynbio. The TourSynbio large model opens up the protein design process and realizes "Protein Design AI in One". It can provide in-depth representation of any protein, support natural language dialogue and prompts, and greatly simplify the protein design process.
August 2024Zhang Haicang's team from the Institute of Computing Technology of the Chinese Academy of Sciences proposed CarbonNovo.This achievement was published in ICML2024. CarbonNovo jointly designs the protein backbone structure and sequence in an end-to-end manner. It effectively improves the design efficiency and performance by establishing a joint energy model and introducing a protein language model, showing significant advantages over the existing two-stage design model.
Paper link:
https://openreview.net/pdf?id=FSxTEvuFa7 Code link:
https://github.com/zhanghaicang/carbonmatrix_public
In October 2024, Professor Liu Haiyan and Professor Chen Quan's team from the School of Life Sciences and Medicine of USTC,We developed a protein backbone denoising diffusion probability model SCUBA-D that does not rely on a pre-trained structure prediction network.It can automatically design the main chain structure from scratch, forming a complete tool chain that can design artificial proteins with new structures and sequences from scratch. It is the only fully experimentally verified protein design method from scratch besides RosettaDesign. Related results have been published in Nature Methods.
Paper link:
https://doi.org/10.1038/s41592-024-02437-W
In 2025, Lu Peilong's team at Westlake University combined deep learning and energy-based methods toThe transmembrane fluorescence-activated protein tmFAP, which can specifically bind to fluorescent ligands, was successfully designed.The deep learning algorithm was used to solve the core problem in transmembrane protein design, and for the first time, the precise de novo design of non-covalent interactions between transmembrane proteins and ligand molecules in the membrane was achieved, and its fluorescence activation ability in living cells was demonstrated, opening up a new path for the design and application of transmembrane proteins. The research has been published in the top international academic journal Nature.
Paper link:
https://www.nature.com/articles/s41586-025-08598-8
At present, China has formed a unique technological ecosystem in the field of AI-driven protein design. Its breakthrough progress is not only reflected in the algorithm innovation level, but also in the construction of a complete innovation chain from basic theory to industrial application. The emergence of these achievements fully demonstrates the depth and breadth of China's technological breakthroughs in the field of protein design. With the continuous development of AI technology, I believe that there will be more remarkable achievements in the future, which will bring about a paradigm shift in the development of global life science research and biopharmaceutical industry.








