Command Palette
Search for a command to run...
University of Florida Uses Neural Networks to Decipher GPCR-G Protein Coupling Selectivity
Contents at a Glance:G protein-coupled receptors (GPCRs) are transmembrane proteins that transmit stimuli outside the cell membrane to the cell membrane and are widely involved in human physiological activities. Recently, researchers at the University of Florida determined the binding selectivity of GPCRs and G proteins, developed an algorithm to predict the selectivity of the two, and studied the structural basis of this selectivity.
Keywords:GPCR neural network drug development
Author | Xuecai
Editor | Sanyang
G protein-coupled receptors (GPCRs) are transmembrane proteins that transmit stimuli outside the cell membrane to the cell membrane. By activating G proteins in the cell membrane and their downstream signaling pathways, GPCRs can be widely involved in important physiological activities such as development, immunity, hormone regulation and neural activity.
G proteins are composed of Gα, Gβ and Gγ subunits, and their diversity determines the diversity of GPCRs signaling responses. The human genome encodes a total of 16 Gα subunits, which are divided into four subfamilies: Gαi/o , Gαq , Gαs and Gα12/13 G proteins determine the downstream signaling pathways and thus the cellular responses. Therefore, the selective binding of GPCRs and G proteins is the key to understanding the signaling system of organisms.
The academic community once believed that GPCRs would only couple with a single G protein, and then functionally divided GPCRs into four types. However, recent researchers have found that most GPCRs couple with multiple G proteins to activate complex cellular responses. The one-to-one classification model is no longer sufficient to describe the coupling relationship between GPCRs and G proteins, and the selectivity mechanism of GPCRs-G proteins is not yet clear.
to this end,Researchers at the University of Florida used kinetic measurements and bioluminescence resonance energy transfer (BRET) technology to determine the guanine nucleotide exchange factor of GPCRs for G proteins to analyze the selective binding of the two. Based on this, the researchers classified GPCRs according to their preference for G proteins and established a coarse-grained model of 124 GPCRs including different mammals. Subsequently, an algorithm for predicting GPCRs-G protein selectivity was developed to study the structural basis of selectivity.Related results have been published inCell Reports".

This result has been published in "Cell Reports"
Paper link:
https://doi.org/10.1016/j.celrep.2023.113173
01 BRET: GPCRs-G protein selective quantification
To quantify GPCRs-G protein selectivity, the researchers used the BRET technique to measure G protein activity in living cells.

BRET technology for real-time detection of G protein activity
Subsequently, the researchers conducted a validation study on cholecystokinin type II receptor (CCKBR). The results of the response amplitude (Amplitude) showed that CCKBR can activate Gαi/o , Gαq , Gα15 and Gα12/13 The G proteins of the family have similar activation levels, but cannot activate Gαs Family of proteins.
The results based on activation rate clearly show that CCKBR has an important effect on Gαq The activation effect of the family is the best, followed by Gαi/o , Gα15 and Gα12/13, which shows that the activation rate-based BRET technology can capture subtle differences between the activities of different G proteins.

Amplitude-based BRET results (C) and activation rate-based BRET results (D)
Accordingly,The researchers measured the selectivity of 124 GPCRs and G proteins as the dataset for this study.

Selectivity measurements of class B GPCRs and G proteins
02 Model Construction: Binary Classification Neural Network
The above results show that the BRET technology based on activation rate can distinguish the selectivity of hundreds of GPCRs and G proteins.The researchers developed a machine learning-based algorithm for predicting class A GPCRs-G protein selectivity.
The algorithm has two tasks:
1. Regarding the coupling of GPCRs, determine whether a certain GPCR can couple with G protein, that is, the amplitude>0%;
2. Regarding the selectivity of GPCRs, it is determined that a certain GPCR-G protein couple can be rapidly activated, that is, the activation rate is >30%.
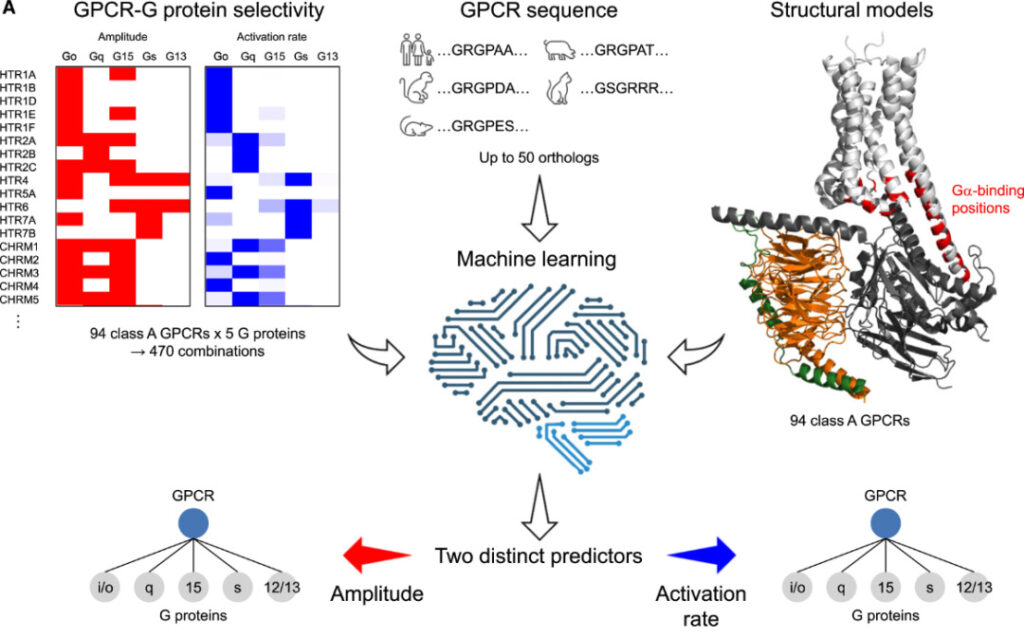
Concept diagram of machine learning algorithms
The coupling of each GPCR and G proteins of different families is a classification problem, so each task can be designed as 5 binary classifications. Based on this, the researchers designed 10 neural network classifiers to handle these tasks. The neural network consists of two fully connected layers (128 and 16 neurons respectively), a flattening layer, three fully connected layers (128, 32 and 4 neurons respectively), and an output layer (1 neuron). The inner layer is activated by a rectified linear unit (ReLU) and finally batch normalized. The output layer is activated by a sigmoid function.
Due to the limited amount of data, 50 homologous sequences were added for each GPCR for data augmentation, assuming that the sequences that determine G protein selectivity are relatively conserved during evolution. The sequence embedding protocol was deployed in the model, and an unsupervised deep learning model was used to describe the properties of protein residues in a specific environment.
The input of the neural network is a tensor of size B*30*1024. The first dimension is the batch size (B=32), the second dimension is the number of residues (30), and the third dimension is eachAmino acid residuesThe size of the pre-trained sequence embeddings (1024).
Averaging of model predictions for amplitude and activation rate AUROC Both are 0.85, indicating that the model has good performance in predicting both indicators.Among them, for Gs The best predictions were for the family proteins, with AUROCs of 0.89 and 0.95, respectively.15 and Gα12/13 family of proteins, the model showed no obvious learning ability.

ROC curves for amplitude (C) and activation rate (D) prediction
03 Decoding the mechanism of GPCRs-Gα protein selectivity
BRET experiments and machine learning provide a solution to decipher the structural basis of GPCRs-G protein selectivity. Based on this, the researchers investigated the available GPCRs-G protein complexes and analyzed 33 class A receptors to find the structure that determines the class A GPCRs-Gα protein selectivity.
The researchers investigated the residue network of GPCRs-Gα proteins and found that all structures of GPCRs facing the cytoplasm were involved in binding to Gα proteins to varying degrees. Similarly, 13 structures in Gα proteins were associated with GPCRs-Gα protein binding, with the C-terminal α-helix (H5) being the most involved.

Interactions between different structural elements of GPCRs and Gα proteins
For the general GPCRs-Gα coupling, GPCRs use ICL2, H8, and most TM residues to connect to Gα protein. Among them, most structures mainly connect to H5, while ICL2 is more extensively connected.
GPCRs and Gαi/o and Gαq The protein coupling patterns of the two families are similar, with the only difference being that the connection of GPCRs to the former is heavily dependent on TM6, while the connection to the latter is not.s Among the connections, the share of ICL2 and ICL3 was greatly reduced, and it was more dependent on TM3 and TM5. The above results show that for different families of Gα proteins, their connection with GPCRs depends on different structures.
Furthermore, by combining the selectivity sequence of GPCRs-G proteins, we investigated the effects of specific structures on Gα proteins of different families.i/o Can the bound GPCRs bind to Gα15 Compared with the former, the connection between GPCRs and the latter cuts off the connection between ICL3 and H4, weakens the interaction between ICL2-H5, and strengthens the connection between TM4-HN and ICL2-s2s3. This suggests that the connection between ICL2 and other residues may be related to Gαi/o or Gα15 Major differences in the linked GPCRs.

GPCRs only interact with Gαi/o Residue network connected to GPCRs and Gα15/Gαi/o Connected residue network (K)
Similarly, GPCRs and Gαs and Gαi/o After combining the residue networks, the results showed that the connection between ICL1 and TM5 was the main difference between the two.
The above results show thatBRET and machine learning can analyze the protein residue network of GPCRs-G protein binding and find the structural basis of their selectivity, providing a new method for the study of GPCRs.
04 AI-GPCR: Unexplored Regions of 96.4%
Over the past decade, the proportion of AI and machine learning applications in the GPCR field has steadily increased. In 2022, 3.6% of GPCR-related papers mentioned AI-related methods.
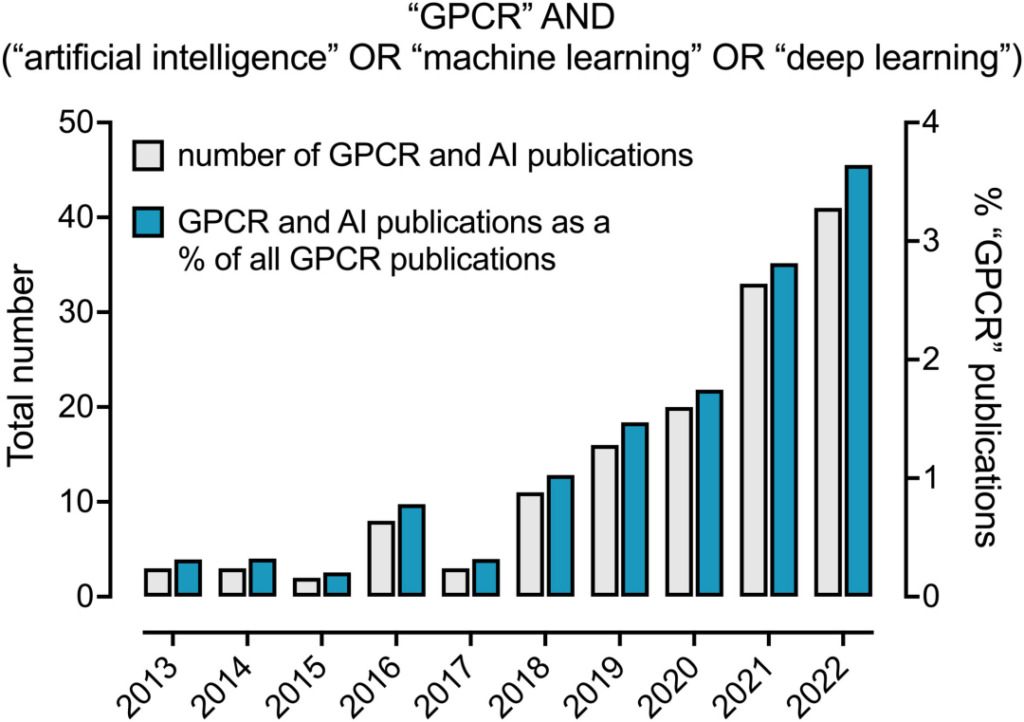
The proportion of AI mentioned in GPCR-related papers
Given the increasing application of AI in GPCR drug research, corresponding algorithms are also being developed. For classification problems, the most commonly used algorithms are those in traditional machine learning, such as those in the scikit-learn library, including support vector machines (SVMs), decision trees, gradient boosting machines, and k-nearest neighbor algorithms.
For numerical results, such as the affinity of protein-ligand binding, regression algorithms are often used to solve them, such as multivariate linear regression, support vector machines, and deep learning networks.
Recent achievements have mostly used deep learning algorithms such as multi-layer perceptrons and convolutional neural networks (CNN) for prediction. With the development of deep learning generation algorithms, protein ligand and structure design has become more efficient and accurate. Generative adversarial networks, recurrent neural networks, reinforcement learning and other algorithms can use the automatic construction of vector space and adaptive metrics to explore larger generation spaces.

The role of AI in each stage of GPCR drug development
Therefore, these algorithms can generate more ligands with desired functions or more accurately predict the structures of unknown proteins, e.g. AlphaFold2 Although models such as AlphaFold2 are not specifically designed to predict the structure of GPCRs, they can still predict the structure of GPCRs efficiently and accurately. In addition, unsupervised or self-supervised deep learning is also emerging in drug discovery.
It can be seen that AI-GPCR may be a new direction for future drug development, but it also leaves us with an unknown area of 96.4%. With the help of efficient classification and accurate prediction algorithms, people can have a clearer understanding of the coupling mechanism of GPCR and inject new momentum into the development of biomedicine.