Command Palette
Search for a command to run...
PaperSearchQA: Lernen des Suchens und Schlussfolgerns in wissenschaftlichen Papieren mit RLVR
PaperSearchQA: Lernen des Suchens und Schlussfolgerns in wissenschaftlichen Papieren mit RLVR
James Burgess Jan N. Hansen Duo Peng Yuhui Zhang Alejandro Lozano Min Woo Sun Emma Lundberg Serena Yeung-Levy
Zusammenfassung
Suchagenten sind Sprachmodelle (LMs), die Schlussfolgerungen ziehen und Wissensbasen (oder das Web) durchsuchen, um Fragen zu beantworten; neuere Methoden optimieren dabei ausschließlich die Genauigkeit der Endantwort mittels Verstärkendem Lernen mit überprüfbarer Belohnung (Reinforcement Learning with Verifiable Rewards, RLVR). Die meisten RLVR-Suchagenten befassen sich mit allgemeinen Domänen im Bereich der Fragen-und-Antwort-Verarbeitung (QA), was ihre Relevanz für technische KI-Systeme in Wissenschaft, Ingenieurwesen und Medizin einschränkt. In dieser Arbeit schlagen wir vor, Agenten zu trainieren, die wissenschaftliche Arbeiten durchsuchen und darin schlussfolgern – dies testet technische Fragen-und-Antwort-Verarbeitung, ist direkt relevant für echte Wissenschaftler und stellt eine entscheidende Fähigkeit für zukünftige KI-Wissenschaftler-Systeme dar. Konkret veröffentlichen wir eine Suchkorpus von 16 Millionen biomedizinischen Kurzreferaten und erstellen eine anspruchsvolle Fakten-QA-Datensammlung namens PaperSearchQA mit 60.000 Fragen, die aus dem Korpus beantwortbar sind, sowie entsprechende Benchmarks. Wir trainieren Suchagenten in dieser Umgebung, um nicht-RL-basierte Retrieval-Baselines zu übertreffen; außerdem führen wir eine detaillierte quantitative Analyse durch und beobachten interessante Agentenverhalten wie Planung, Schlussfolgerung und Selbst-Verifikation. Unser Korpus, unsere Datensätze und Benchmarks sind mit dem populären Search-R1-Codebase für RLVR-Training nutzbar und sind unter https://huggingface.co/collections/jmhb/papersearchqa veröffentlicht. Schließlich sind unsere Methoden zur Datenerstellung skalierbar und leicht auf andere wissenschaftliche Domänen übertragbar.
One-sentence Summary
Stanford and Chan Zuckerberg Biohub researchers propose PaperSearchQA, a 60k-sample biomedical QA dataset with 16M abstracts, enabling RLVR-trained search agents to outperform retrieval baselines via planning and self-verification—advancing AI Scientist systems for technical domains.
Key Contributions
- We introduce a new scientific QA environment for training search agents, featuring a corpus of 16 million biomedical abstracts and a 60k-sample factoid dataset (PaperSearchQA), designed to test technical reasoning and support real-world scientific workflows.
- We demonstrate that RLVR-trained agents outperform non-RL baselines on this task, while also revealing emergent behaviors such as planning, self-verification, and strategic query rewriting through quantitative and qualitative analysis.
- Our datasets and benchmarks are compatible with the Search-R1 codebase and publicly released on Hugging Face, with scalable data construction methods extendable to other scientific domains like chemistry or materials science.
Introduction
The authors leverage reinforcement learning with verifiable rewards (RLVR) to train language models that search and reason over scientific papers — a capability critical for AI systems in science, engineering, and medicine. Prior RLVR work focused on general-domain trivia, which lacks the technical depth needed for real scientific workflows. Existing scientific QA systems rely on scaffolding or supervised fine-tuning, limiting their ability to generalize. The authors’ main contribution is a new training environment including a 16M-abstract biomedical corpus, a 60k-sample factoid QA dataset (PaperSearchQA), and benchmarks — all compatible with the Search-R1 codebase — enabling RL-trained agents to outperform non-RL baselines and exhibit emergent behaviors like planning and self-verification.
Dataset
-
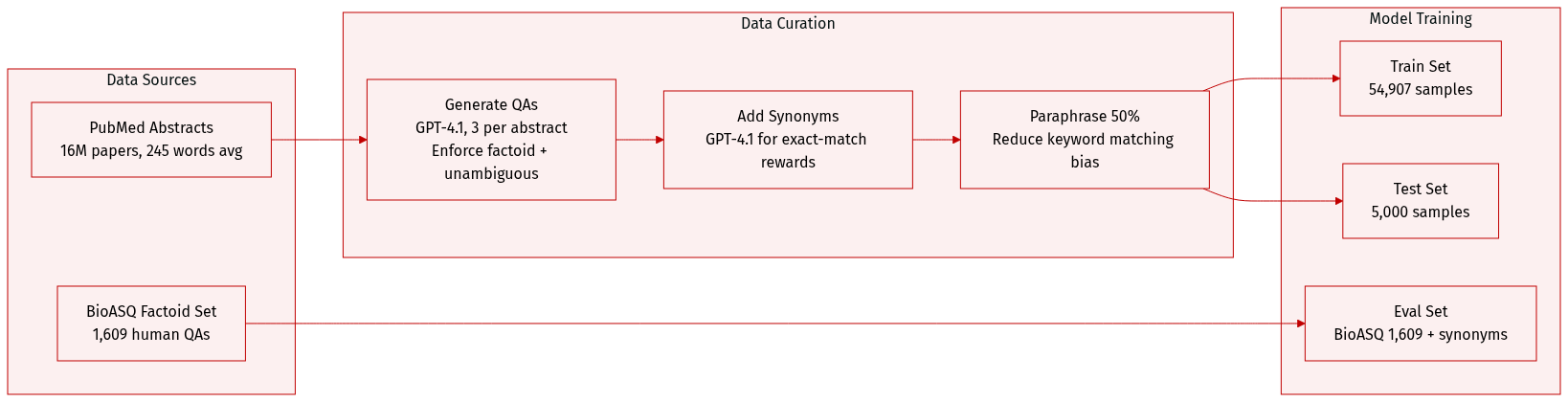
The authors use PaperSearchQA, a biomedical QA dataset built from 16M PubMed abstracts, to train and evaluate retrieval-augmented agents. It contains 54,907 training and 5,000 test samples, each with a factoid answer, category label, and paraphrase flag.
-
Questions are generated via GPT-4.1 using a structured prompt that enforces unambiguous, single-entity answers and avoids acronyms or document-specific phrasing. Each abstract yields 3 QAs. Half are paraphrased via LLM to reduce keyword matching bias.
-
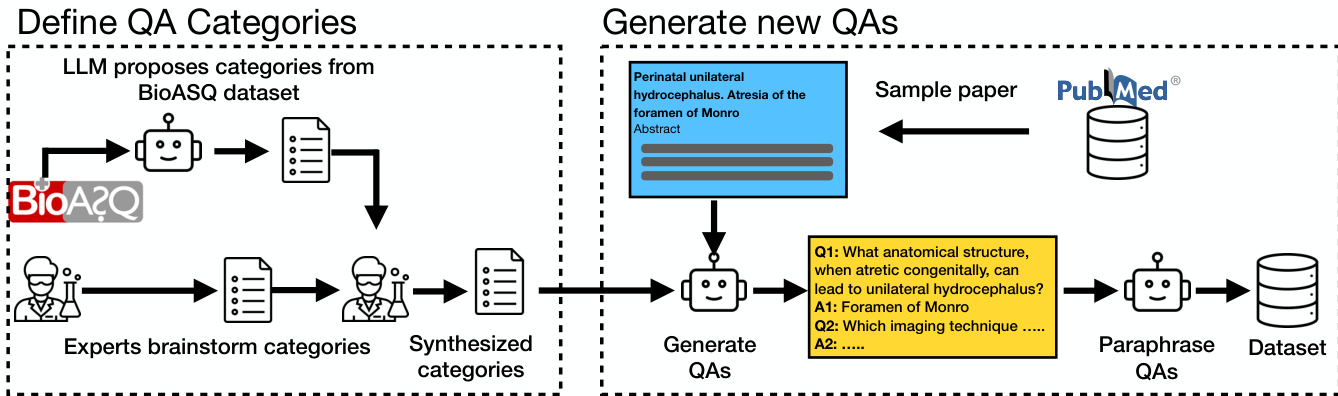
Ten expert-defined categories guide QA generation, including “Experimental & computational methods” (27%) and “Therapeutics, indications & clinical evidence.” Categories were derived by synthesizing human brainstorming and LLM analysis of BioASQ questions.
-
Synonyms for each ground truth answer are generated using GPT-4.1 to support exact-match reward modeling. All samples include PubMed ID, category, and paraphrase status. The dataset is CC-BY-4.0 licensed and available on Hugging Face.
-
For evaluation, the authors use BioASQ’s factoid subset (1,609 samples), which they augment with answer synonyms using the same LLM method. BioASQ questions are human-written and cover broader question types, but only factoid QAs are used in this work.
-
The retrieval corpus consists of 16M PubMed abstracts (mean 245 words), indexed with BM25 (2.6GB) and e5 (93GB). At inference, e5 requires two A100 GPUs. The corpus and BioASQ data inherit CC-BY-2.5 licensing from BioASQ.
-
The full pipeline cost ~$600 via OpenRouter. Prompts and code are publicly available; the dataset is designed for RLVR training where reward models verify exact match or synonym match without requiring reasoning traces or retrieved document annotations.
Method
The authors leverage a reinforcement learning framework with verifiable rewards (RLVR) to train search-capable language models, enabling agents to iteratively reason, issue search queries, and synthesize answers based on retrieved evidence. The training pipeline begins with constructing domain-specific QA datasets, followed by indexing a search corpus, and culminates in policy optimization via RLVR. The core interaction loop is governed by a minimal system prompt that instructs the model to encapsulate reasoning within -thinking- tokens, issue search queries via tags, and deliver final answers within tags. This design intentionally avoids prescribing detailed reasoning strategies, allowing the agent to discover effective behaviors through reward-driven exploration.
Refer to the framework diagram, which illustrates a typical agent trajectory: the model first performs internal reasoning to identify key components of the question, then issues a search query to retrieve relevant documents, integrates the retrieved information into its reasoning trace, and finally produces a verified answer. The retrieved documents are appended to the context but excluded from gradient computation during training, ensuring the policy learns to generate useful queries and synthesize answers rather than memorize retrieval outputs.
The training objective maximizes the expected reward over trajectories generated by the policy LLM, πθ, conditioned on a retriever R and a QA dataset D:
πθmaxEx∼D,y∼πθ(⋅∣x;R)[rϕ(x,y)]−βDKL[πθ(y∣x;R)∣∣πref(y∣x;R)]Here, the reward model rϕ(x,y) extracts the final answer from the generated sequence and assigns a binary reward (1 if correct, 0 otherwise). The KL penalty term prevents excessive deviation from the reference policy, which is initialized to the pre-trained LLM state. Optimization is performed using Group Relative Policy Optimization (GRPO), which computes advantages within groups of rollouts to stabilize training. The GRPO objective incorporates clipping and group-normalized advantages to reduce variance and improve sample efficiency.
As shown in the figure below, the training data generation process involves two stages: first, the LLM proposes QA categories from an existing dataset (BioASQ), which are then refined by domain experts; second, the model generates new QA pairs from sampled scientific papers, which are paraphrased and stored in the final dataset. This synthetic data pipeline ensures coverage of diverse scientific domains while maintaining factual grounding.
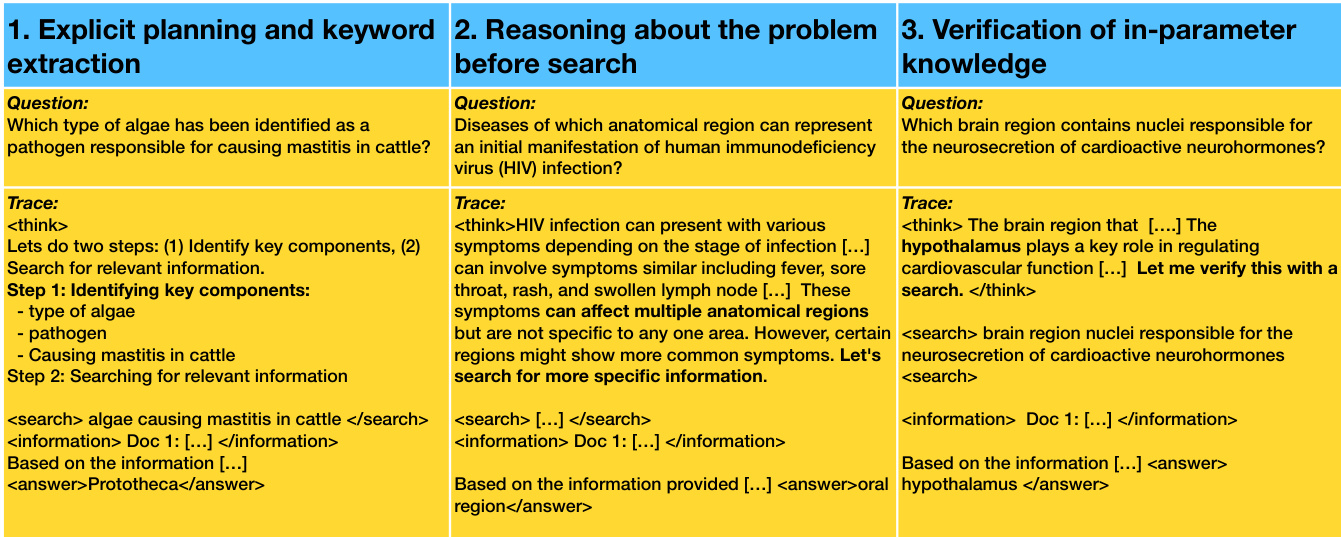
The agent’s behavior during training is characterized by three distinct reasoning modes: explicit planning and keyword extraction, reasoning before search, and verification of in-parameter knowledge. In the first mode, the agent decomposes the question into subtasks and issues targeted search queries. In the second, it performs preliminary reasoning to narrow the scope before retrieving external information. In the third, it leverages internal knowledge to hypothesize an answer and uses search only for verification. These modes are not hard-coded but emerge from the RLVR training process, as the agent learns to allocate effort between internal reasoning and external retrieval based on reward feedback.
Experiment
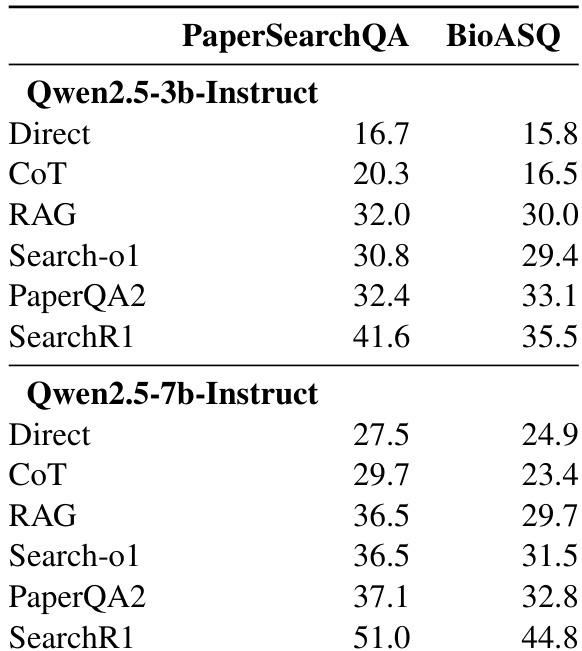
- RLVR training significantly boosts performance on scientific QA tasks compared to baseline methods like direct inference, chain-of-thought, and RAG, especially for factoid questions.
- RLVR-trained models (Search-R1) outperform RAG by 9.6–14.5 points on PaperSearchQA and 5.5–9.3 points on BioASQ, with gains increasing with model size.
- Retrieval method (BM25 vs e5) shows minimal performance difference, suggesting keyword-based retrieval suffices in scientific domains due to technical terminology.
- LLMs retain substantial parametric knowledge of scientific facts, but retrieval remains essential as memorization is incomplete.
- Paraphrasing questions during dataset construction increases difficulty and better tests generalization, as non-paraphrased questions are easier to answer.
- Training dynamics mirror general QA settings; base models require more time to converge and are more stable than instruct models under GRPO.
- Qualitatively, trained agents favor explicit keyword extraction and search planning; early reasoning before search and verification of known answers also occur but diminish with training.
- After retrieval, models typically answer immediately with little explicit reasoning, possibly due to simplified comprehension needs or RL-induced parameter tuning.
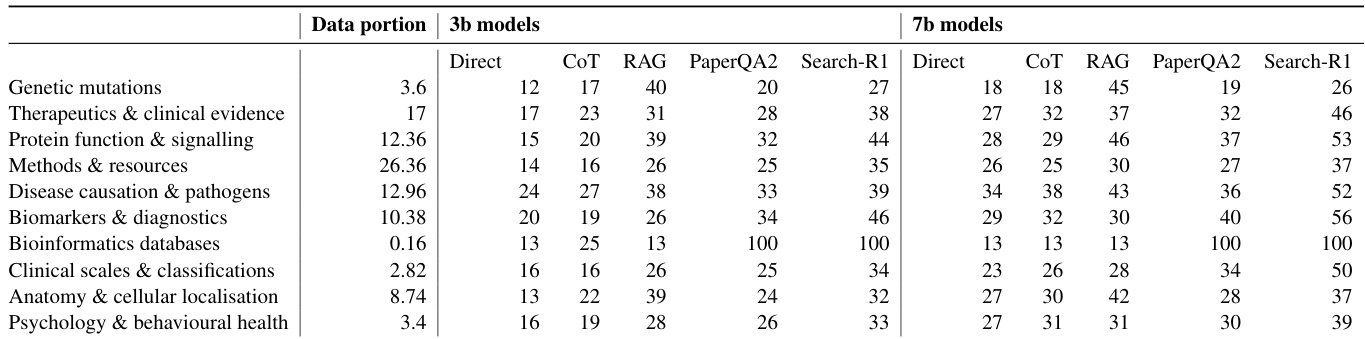
- Performance varies by category, with “Biomarkers & diagnostics” and “Protein function & signalling” being easiest and “Genetic mutations” most challenging.
The authors use reinforcement learning with verifiable rewards (RLVR) to train LLMs on scientific question answering, and results show this approach consistently outperforms baseline methods including direct inference, chain-of-thought, and retrieval-augmented generation. Performance gains are more pronounced with larger models, and the method proves effective across both in-domain and external benchmarks. The improvements suggest RLVR enhances the model’s ability to leverage parametric knowledge rather than relying solely on retrieval or reasoning scaffolds.
The authors use RLVR training to improve LLM performance on scientific question answering, showing consistent gains over baseline methods like RAG and chain-of-thought across both 3B and 7B models. Results show that retrieval-augmented approaches significantly outperform retrieval-free ones, and model size correlates with better performance, suggesting parametric knowledge plays a key role. Per-category analysis reveals that performance varies by domain, with “Bioinformatics databases” being easiest and “Genetic mutations” most challenging, while RLVR training consistently delivers the strongest results across categories.