Command Palette
Search for a command to run...
VenusFactory Protein Engineering Design Platform
Date
Size
6.42 GB
License
Apache 2.0
GitHub
Paper URL

1. Tutorial Introduction

VenusFactory was developed in 2025 by a joint team from Shanghai Jiao Tong University, Shanghai Artificial Intelligence Laboratory, and East China University of Science and Technology. The related research papers are as follows: VenusFactory: A Unified Platform for Protein Engineering Data Retrieval and Language Model Fine-Tuning .
VenusFactory is a unified platform designed specifically for the protein engineering community, aiming to integrate biological data retrieval, standardized task benchmarking, and modular fine-tuning of pre-trained protein language models (PLMs).
The platform supports command-line execution and a Gradio-based code-free interface, and integrates more than 40 protein-related datasets and more than 40 popular PLMs, making it easy for researchers in computer science and biology to use.
The tutorial provides 7 functional modules:
- Training: Zero-code model training, supports 40+ large models, and uses private datasets to train your own models.
- Evaluation: An easy-to-use tool for comprehensive performance evaluation of protein models.
- Prediction: Use the trained model to predict the function of new protein sequences.
- VenusAgent: A protein engineering agent that works with DeepSeek to enable AI protein computation.
- Quick Tools: Easy-to-use version, supports zero-sample mutation prediction (directed evolution) and supervised prediction (function or property prediction).
- Advanced Tools: Advanced customized version, supporting zero-sample mutation prediction (directed evolution) and supervised prediction (function or property prediction).
- Download: Easily link to protein data and support multi-threaded downloads from major databases (RCSB, UniProt...).
The computing resources used in this tutorial are a single RTX 4090 card. The model used in this tutorial is saved in
/openbayes/input/input1All data are stored in the directory/openbayes/home/VenusFactorydirectory.
2. Operation steps
1. Start the container
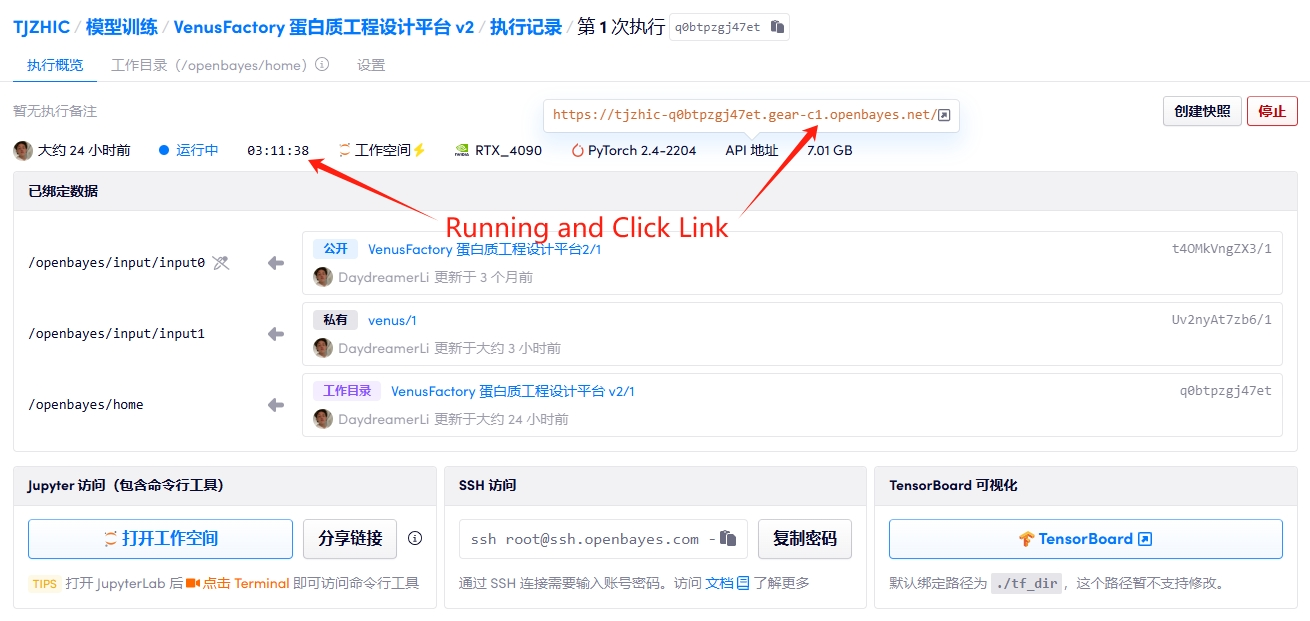
2. Manual
If "Bad Gateway" is displayed, it means the project is initializing. Please wait about 1-2 minutes and refresh the page.
The "Manual" user guide of VenusFactory currently includes four modules: Training, Evaluation, Prediction, and Download.
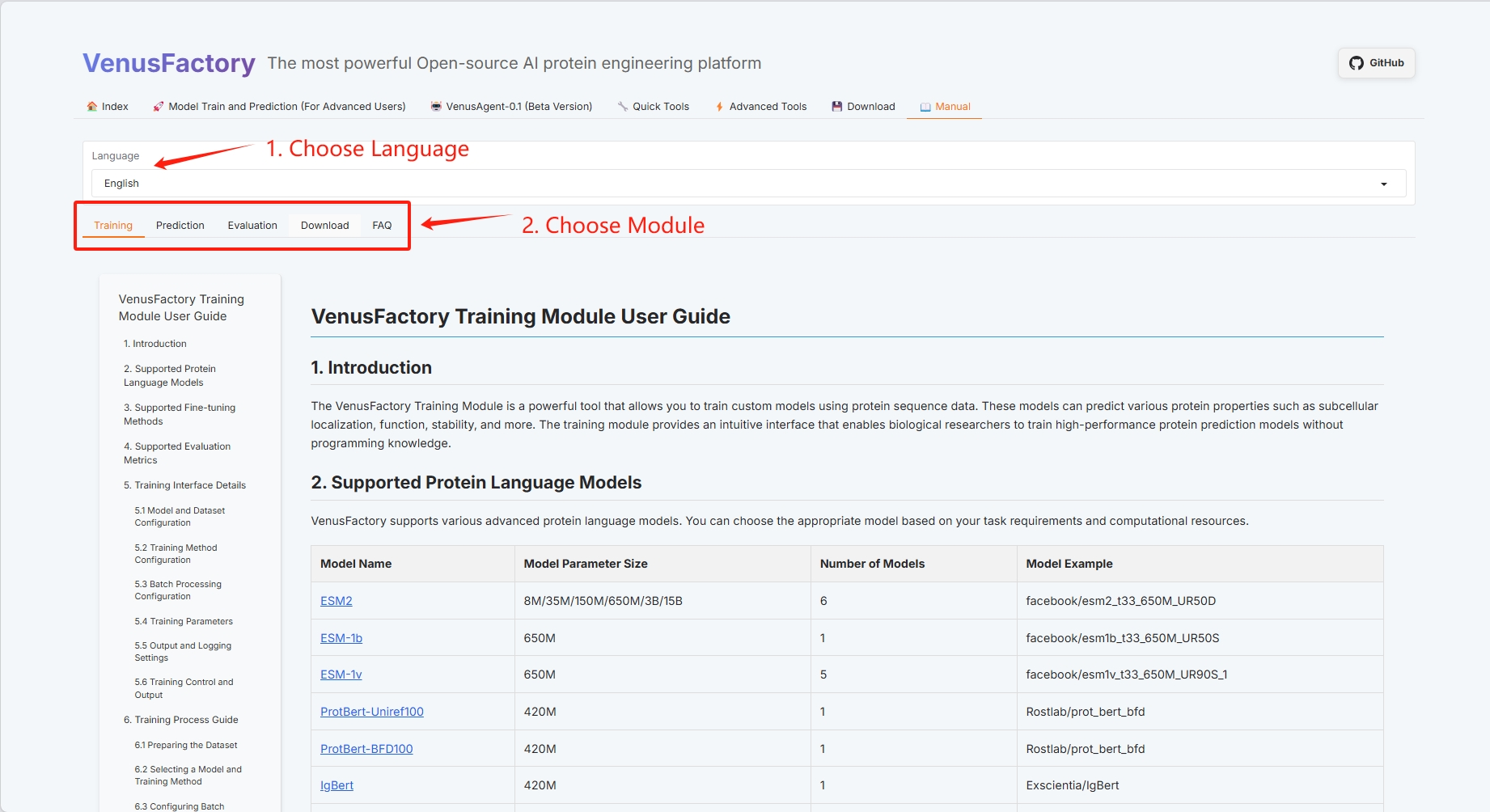
3. Specific function demonstration
3.1 Training
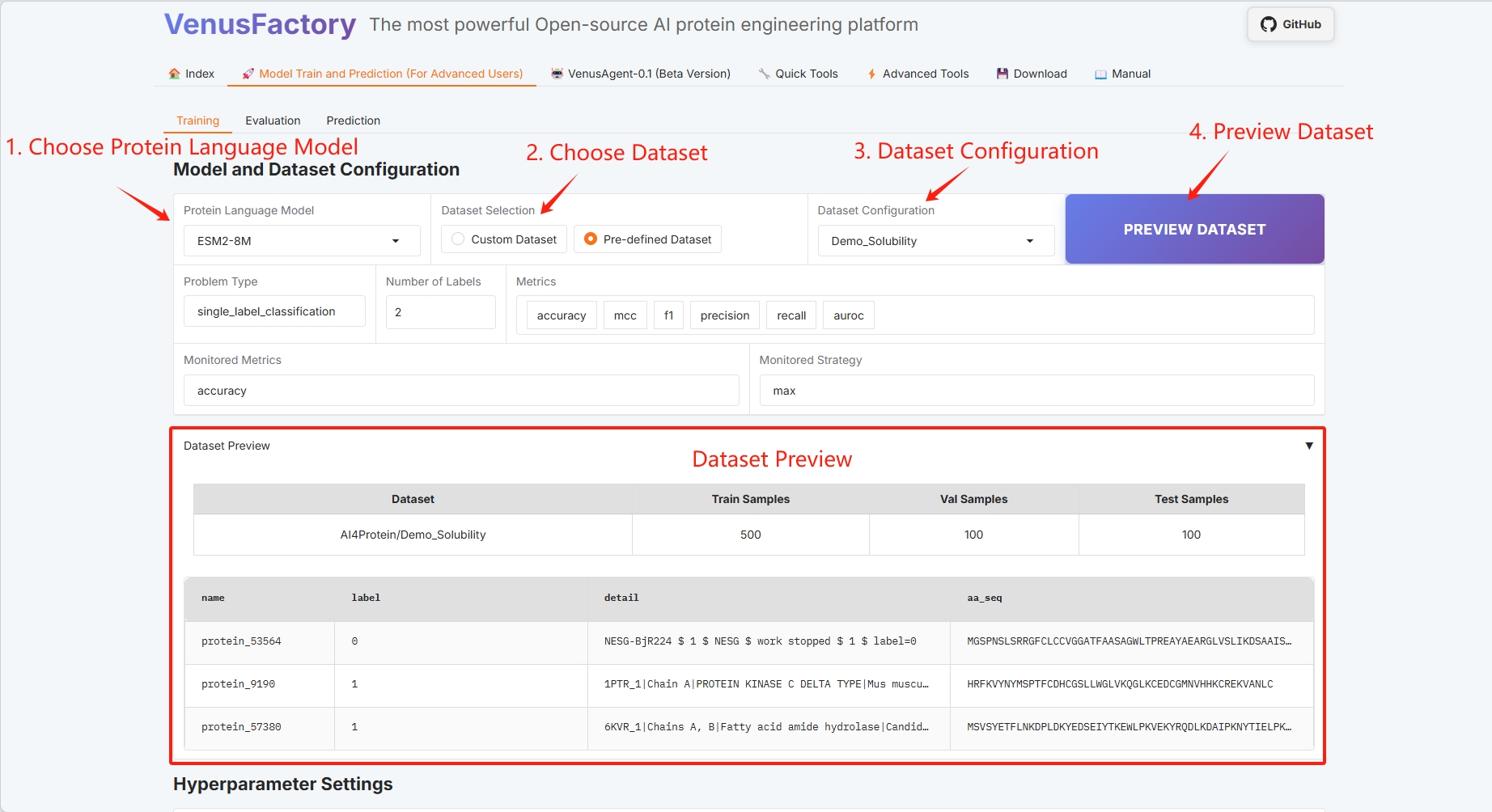
Click the "Training" module in the "Model Train and Prediction Training" module
- Select Protein Language Model
- Dataset selection
- Dataset Preview
- Training method configuration (refer to the user guide for specific information)
- Batch configuration (see the User Guide for details)
If the selected model parameters are large, please replace the graphics card with a larger one.
Set the training model save path and click "START TRAINING" to start training.
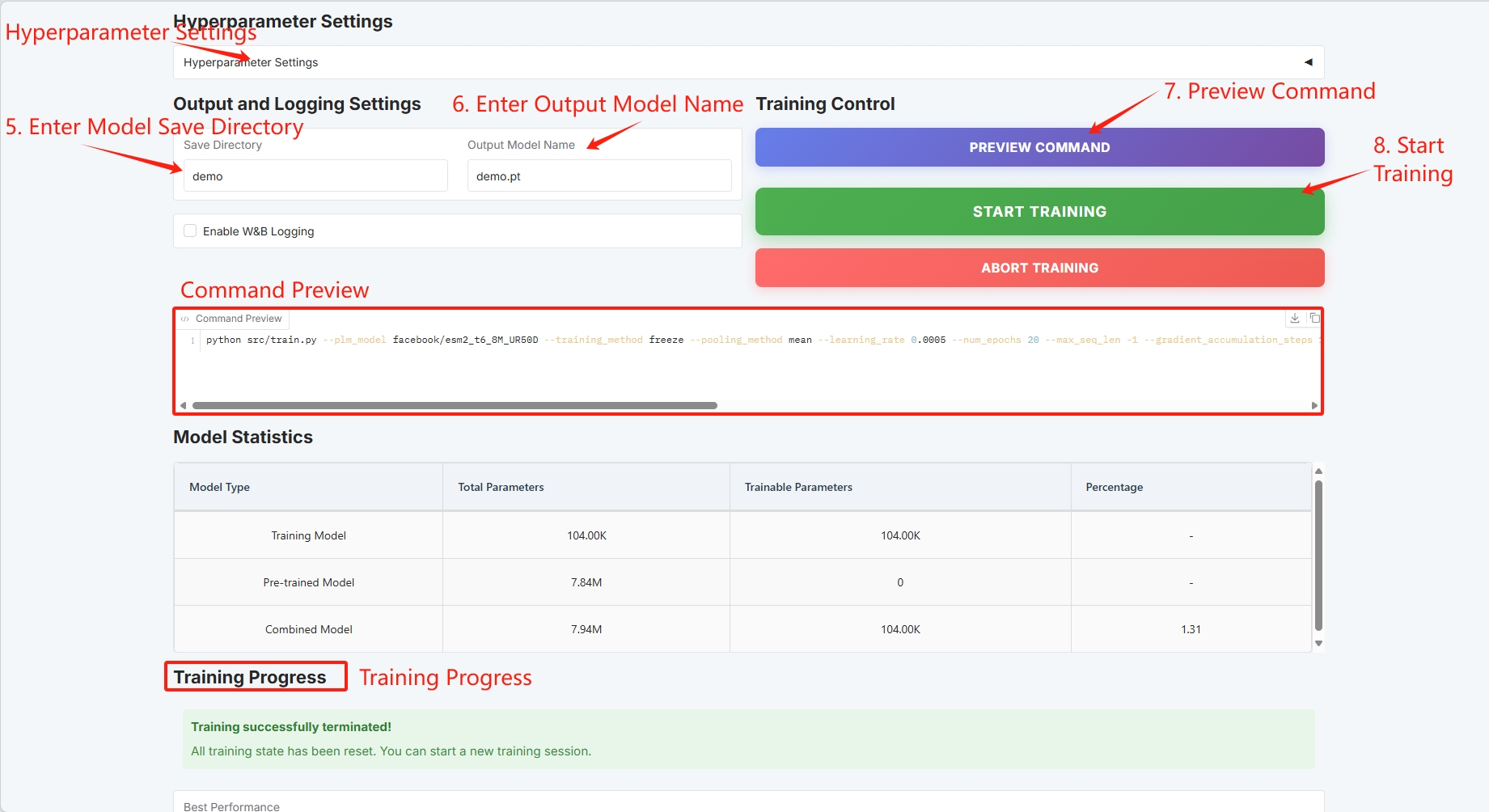
At this point you can see the training parameters and loss curve
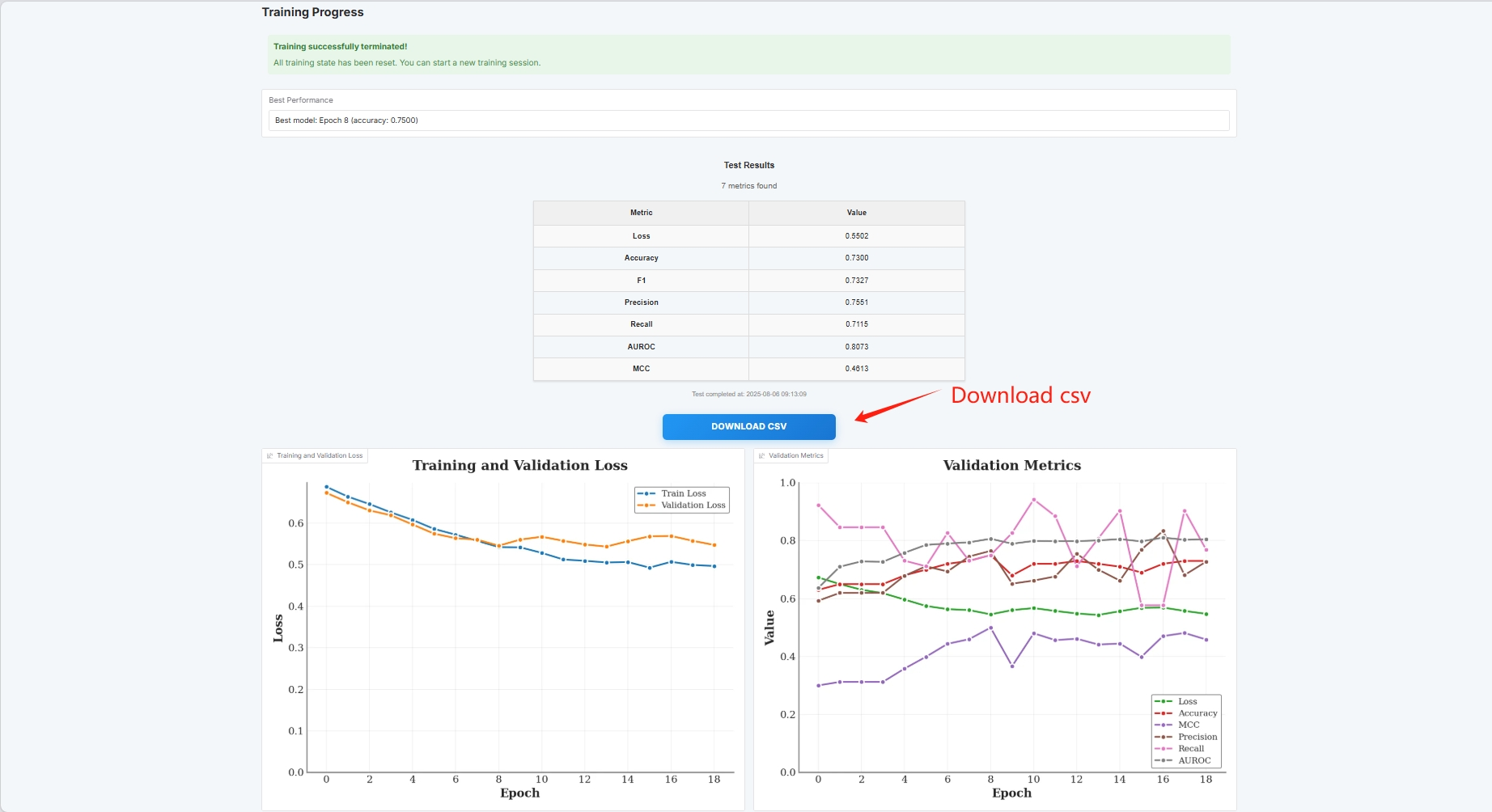
If you want to use your own dataset, you can use the Custom Dataset configuration. Just fill in the path of your dataset (see the Manual documentation for details).
3.2 Evaluation
Click the "Evaluation" module in the "Model Train and Prediction Training" module
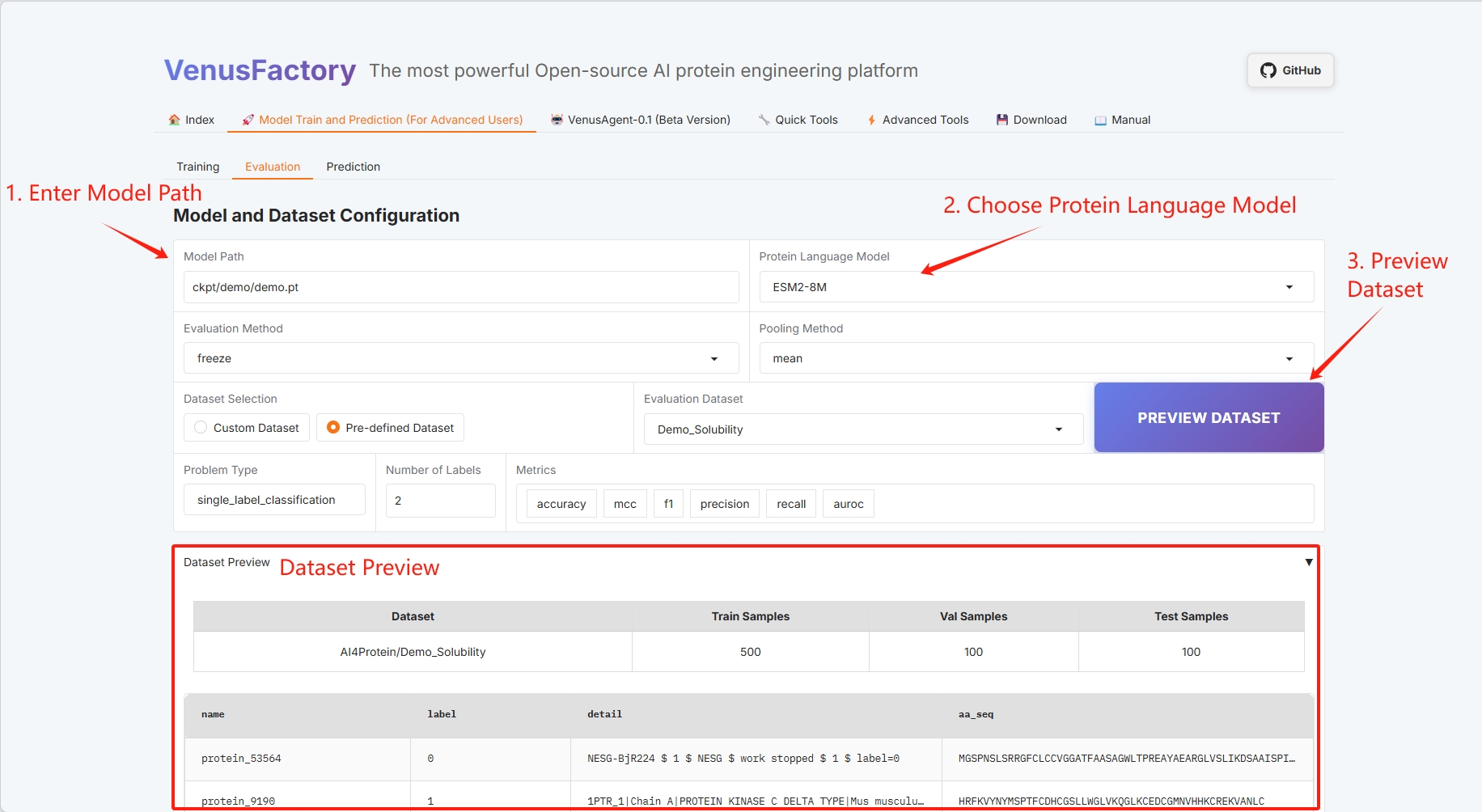
- Model path and protein language model selection
- Evaluation method and pooling method (refer to the user guide for specific information)
- Dataset selection
- Dataset Preview
- Question types and tags (see the User Guide for details)
- Batch configuration (see the User Guide for details)
Set the path to save the trained model and select the protein language model.
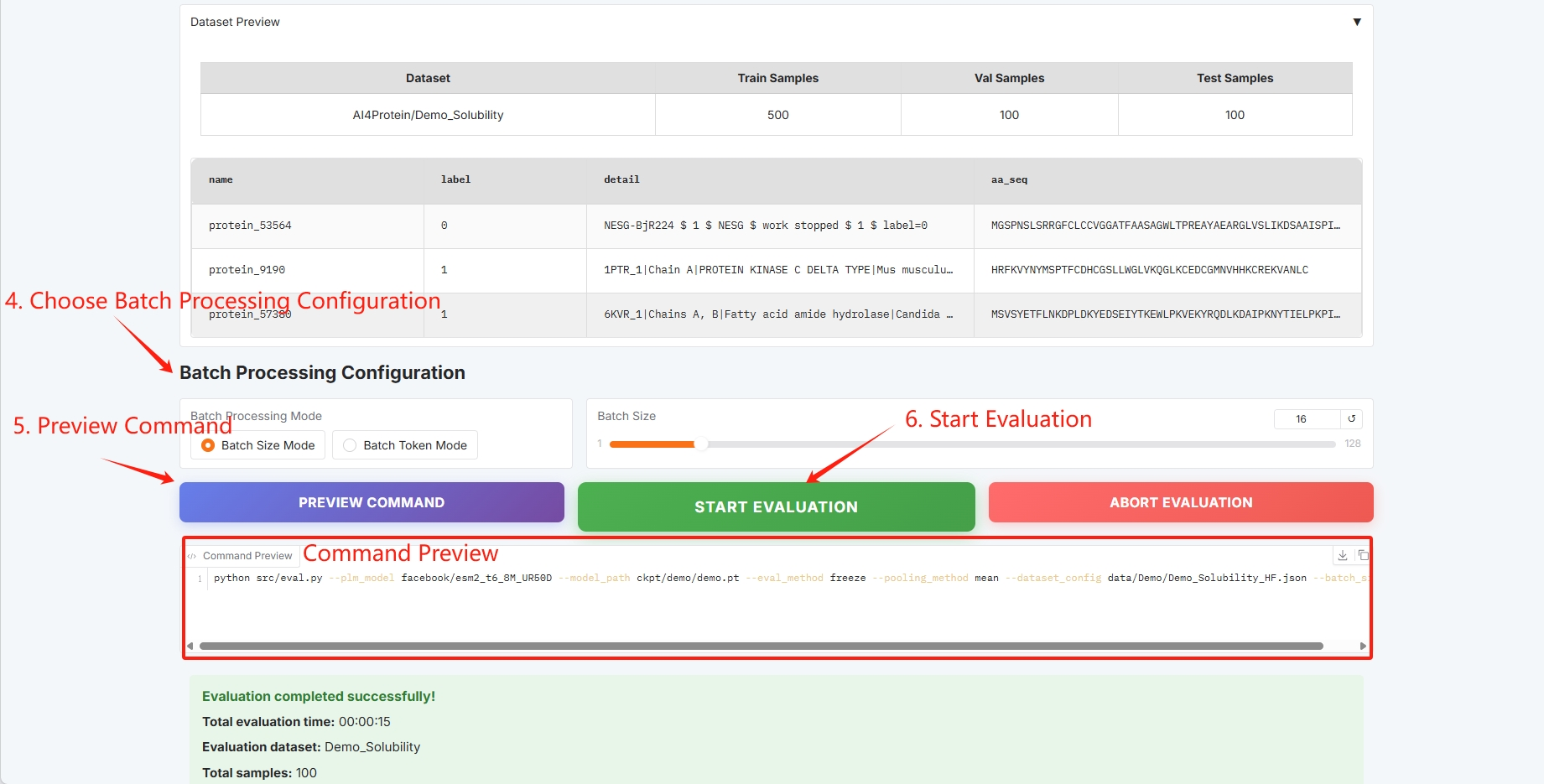
Batch configuration, click "START EVALUATION" to start training.
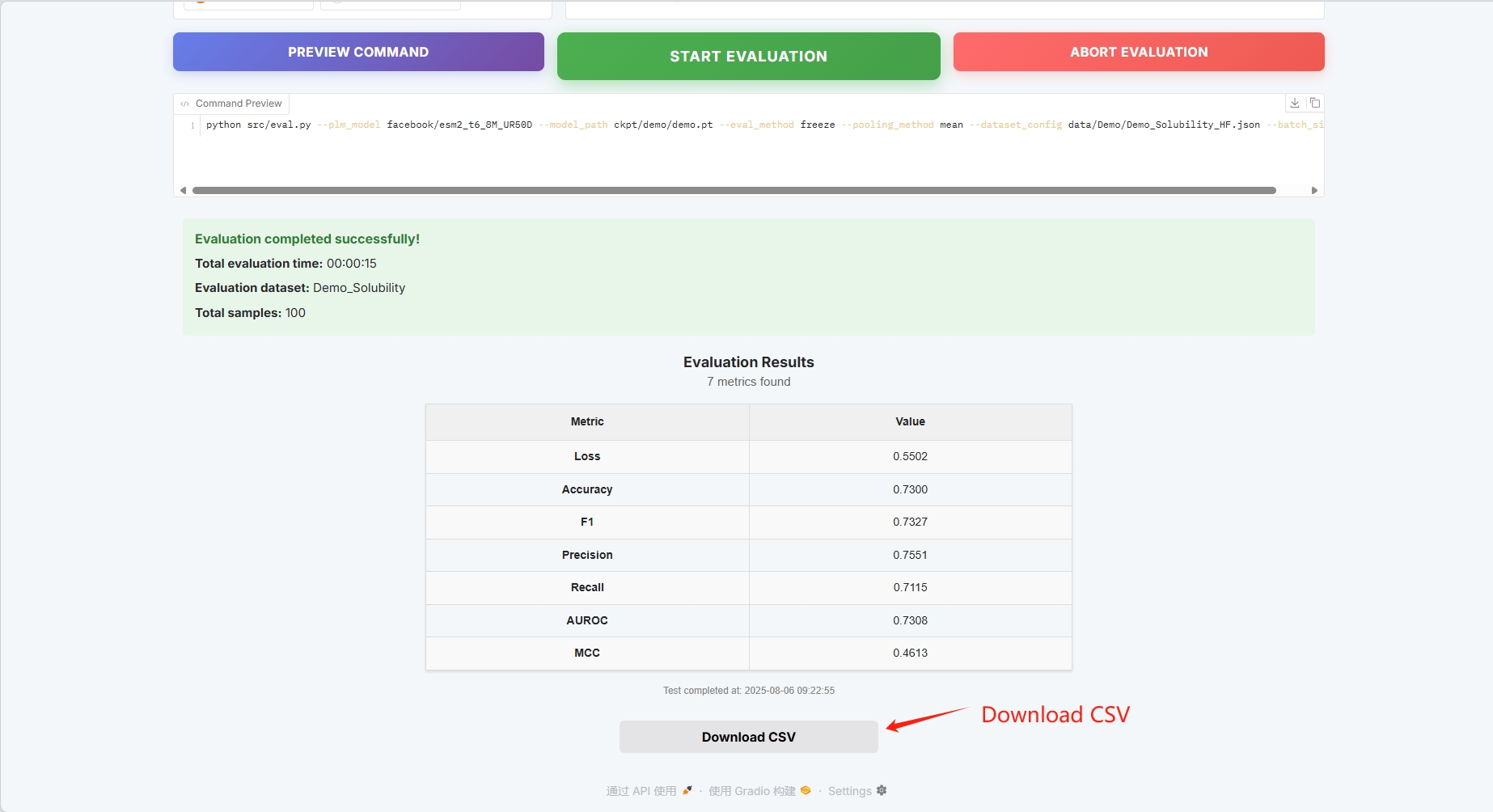
The evaluation results are as follows and can be downloaded as CSV
If you want to use your own dataset, you can use the Custom Dataset configuration. Just fill in the path of your dataset (see the Manual documentation for details).
3.3 Prediction
Click the "Prediction" module in the "Model Train and Prediction Training" module to perform single sequence prediction and batch prediction.
- Model Configuration
- Select the prediction module (refer to the user guide for details)
Set the training model save path, select the protein language model, and click "START PREDICTION" to start training.
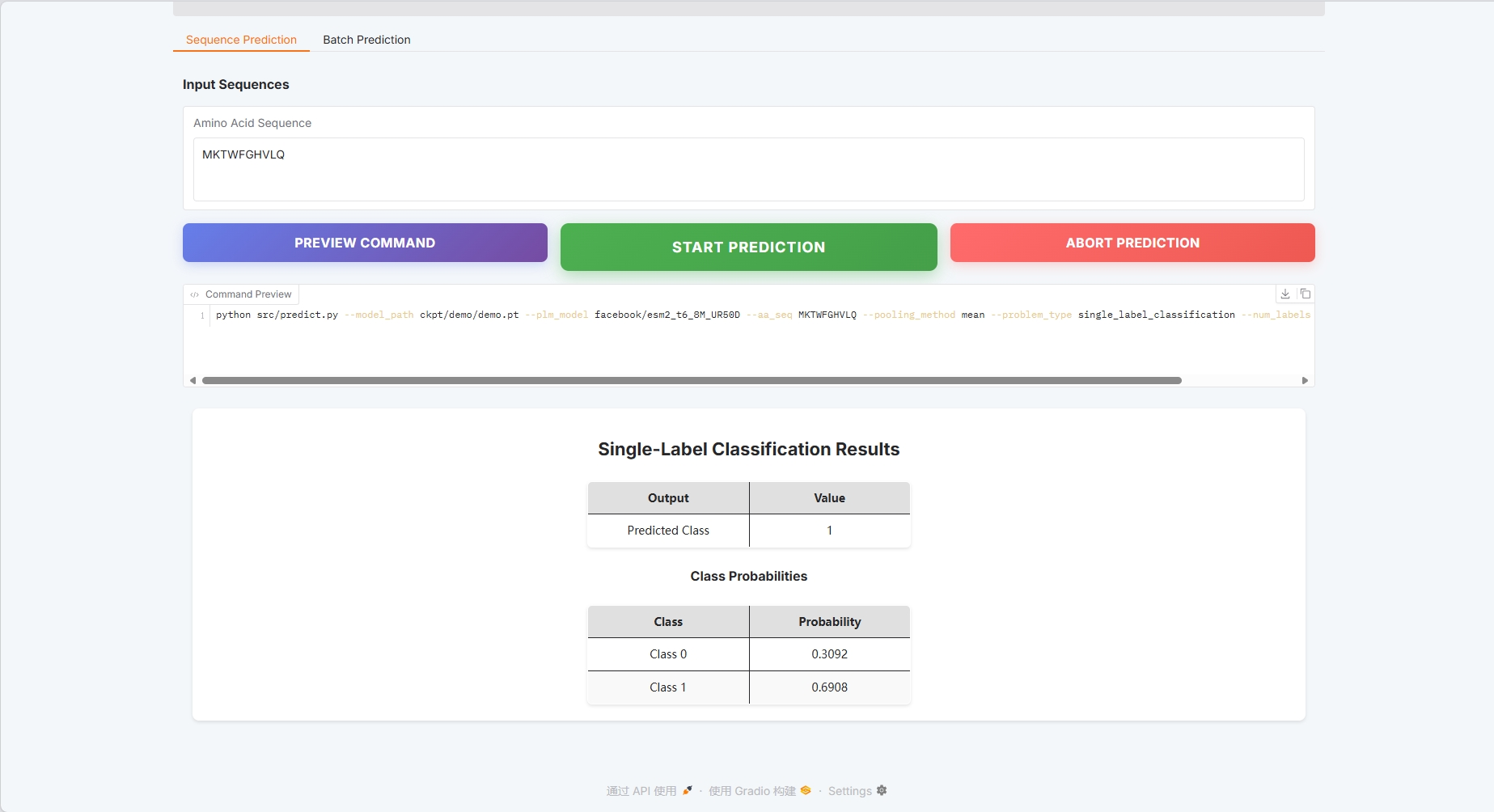
Single sequence prediction
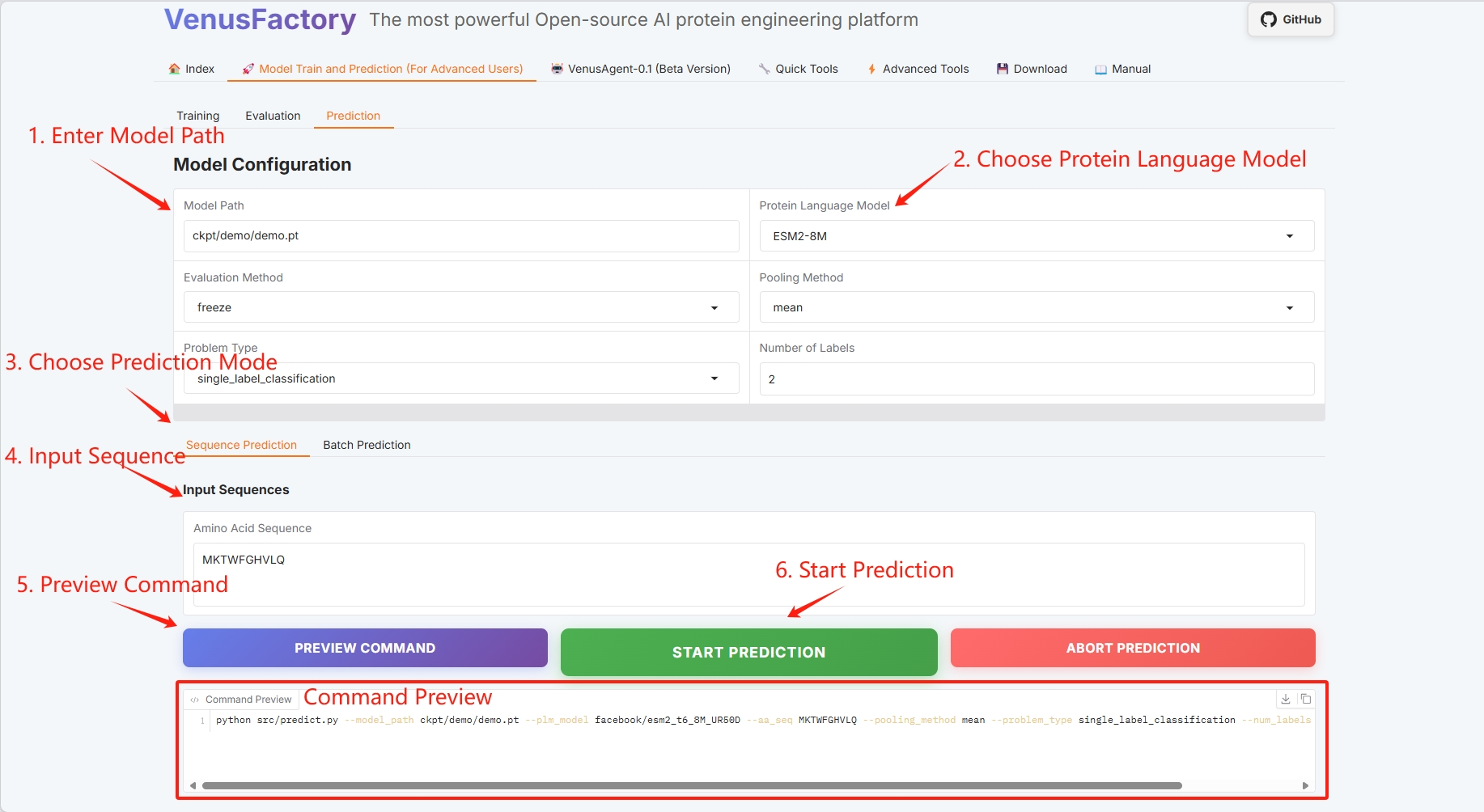
Protein sequence example: MKTWFGHVLQ
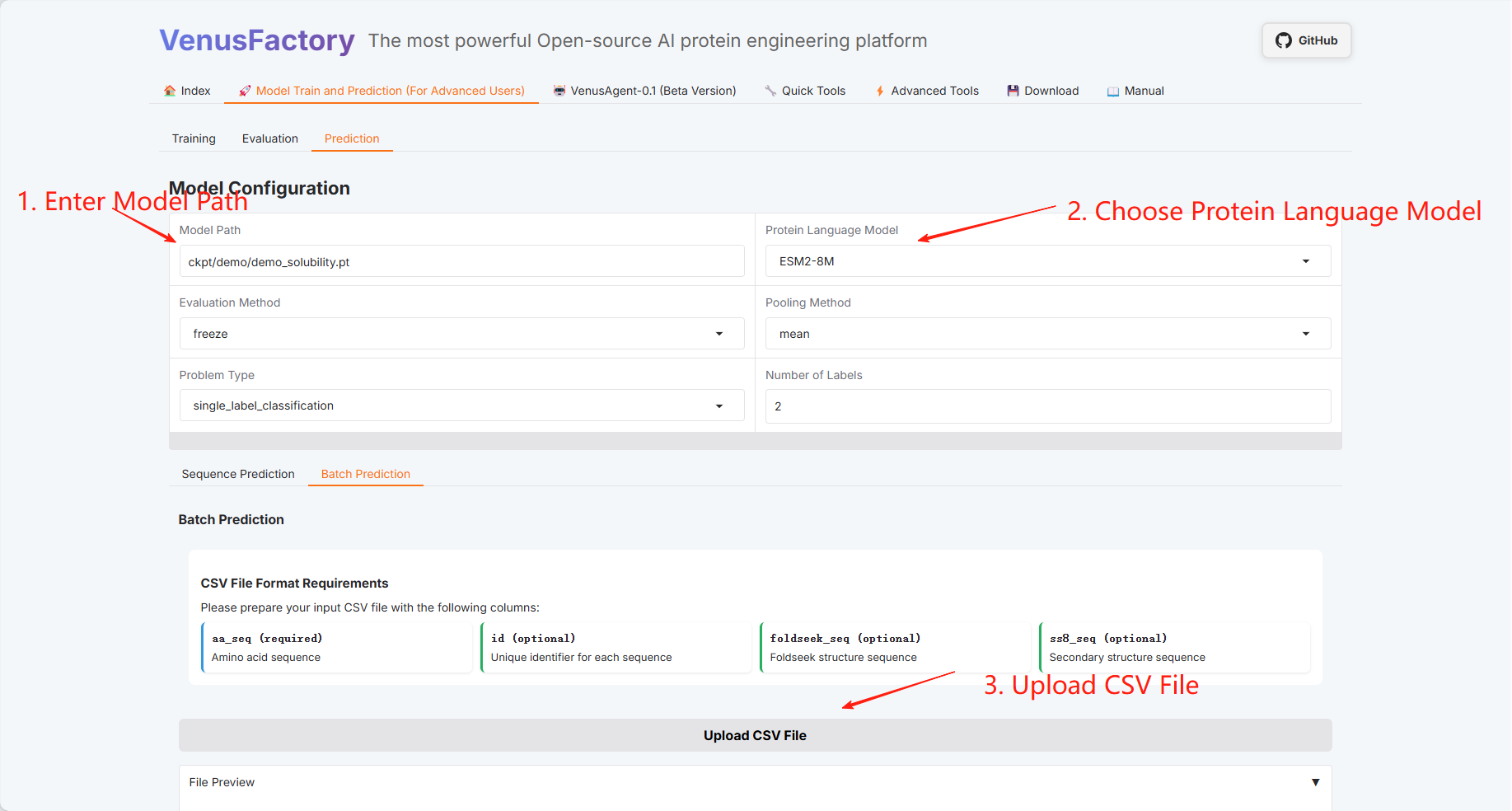
Batch Prediction
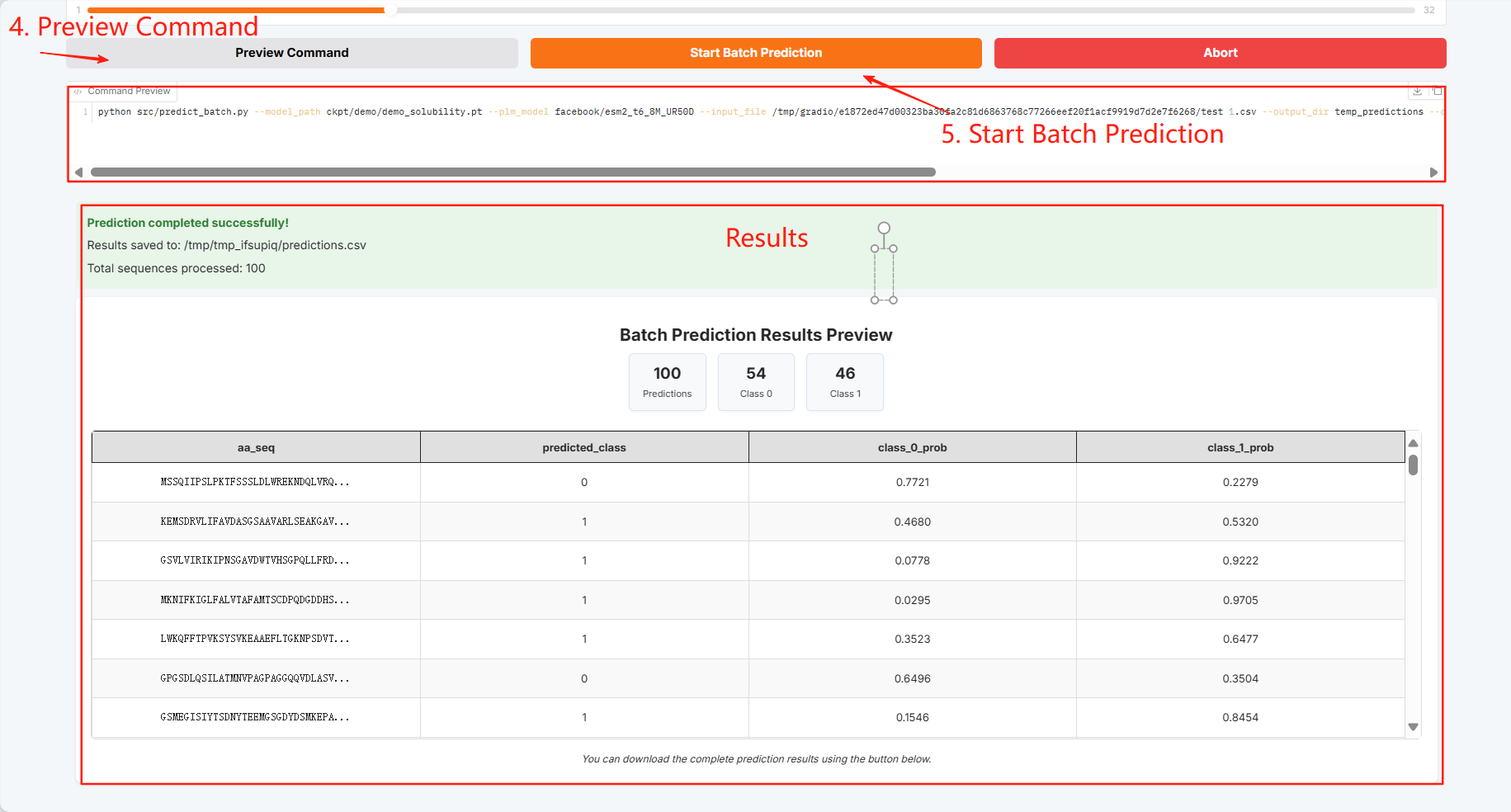
Batch prediction results can be downloaded and saved
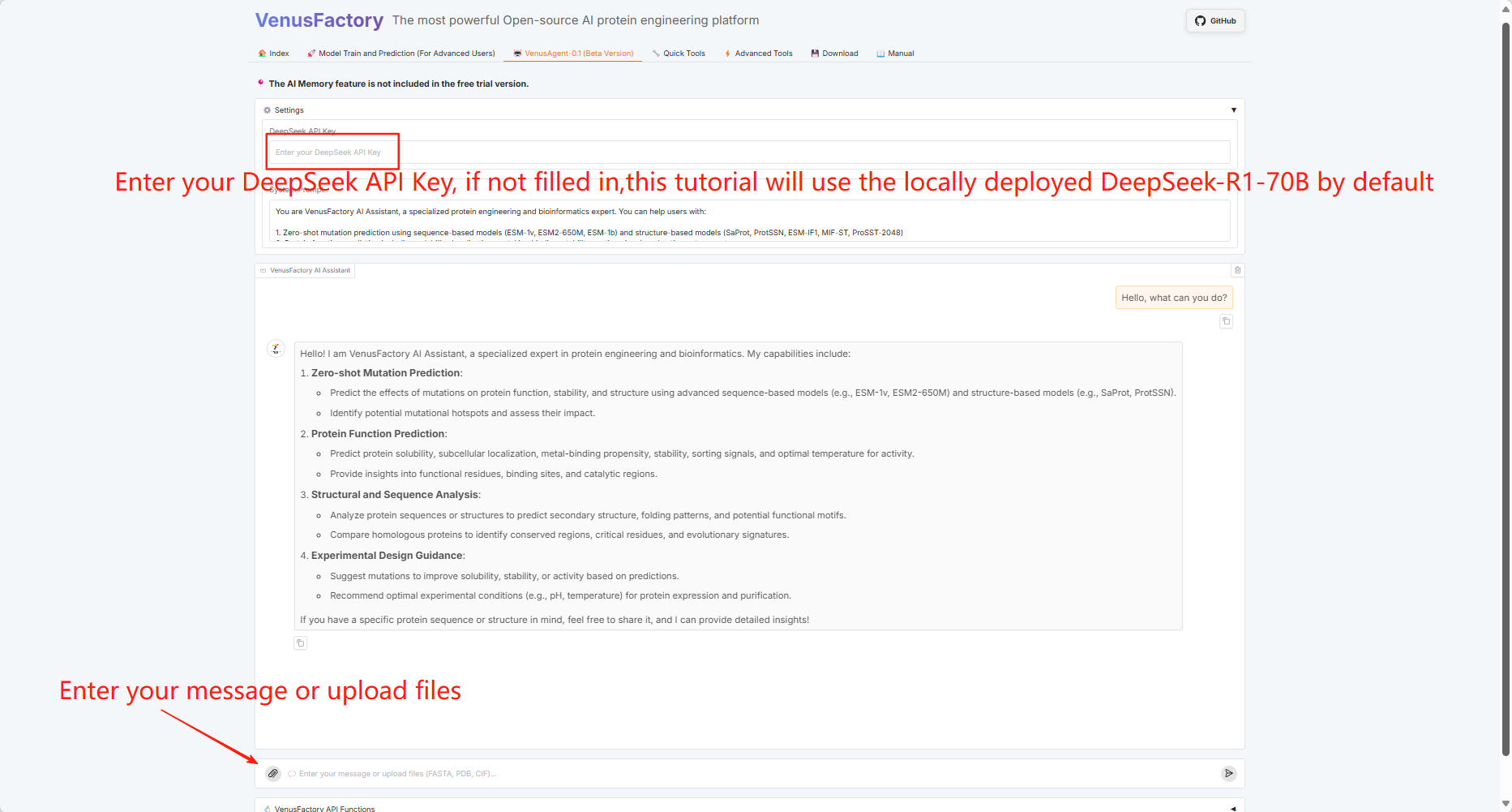
3.4 VenusAgent
Click the "VenusAgent" module
Since VenusAgent needs to call the DeepSeek large model, this tutorial provides two calling methods: enter the API Key yourself or use the DeepSeek-R1-70B model deployed on the platform.
You can choose different graphics card experiences according to the required functions. The card selection instructions are as follows:
If using a single RTX 4090 graphics card, the VenusAgent function does not support the use of locally deployed large model services (using the DeepSeek API Key is unlimited).
If you use dual RTX 4090 graphics cards, you cannot use other functions immediately (after 1-2 minutes) after using the VenusAgent function (there is no restriction when using the DeepSeek API Key).
If you use dual RTX A6000 graphics cards, VenusAgent functions are unlimited.
The computing resources used in this tutorial are a single RTX 4090 card. The model used in this tutorial is saved in
/openbayes/input/input1All data are stored in the directory/openbayes/home/VenusFactorydirectory.
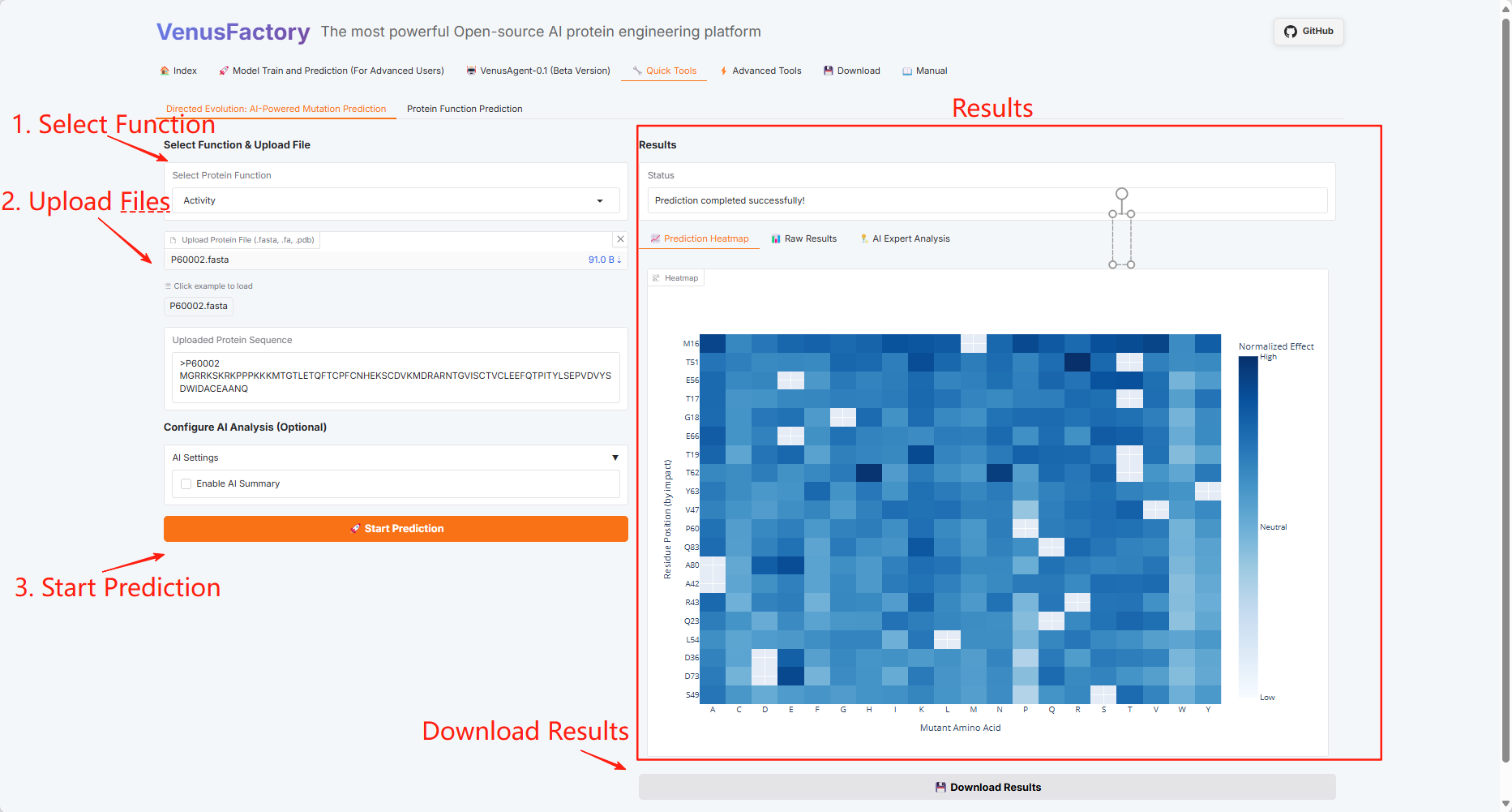
3.5 Quick Tools
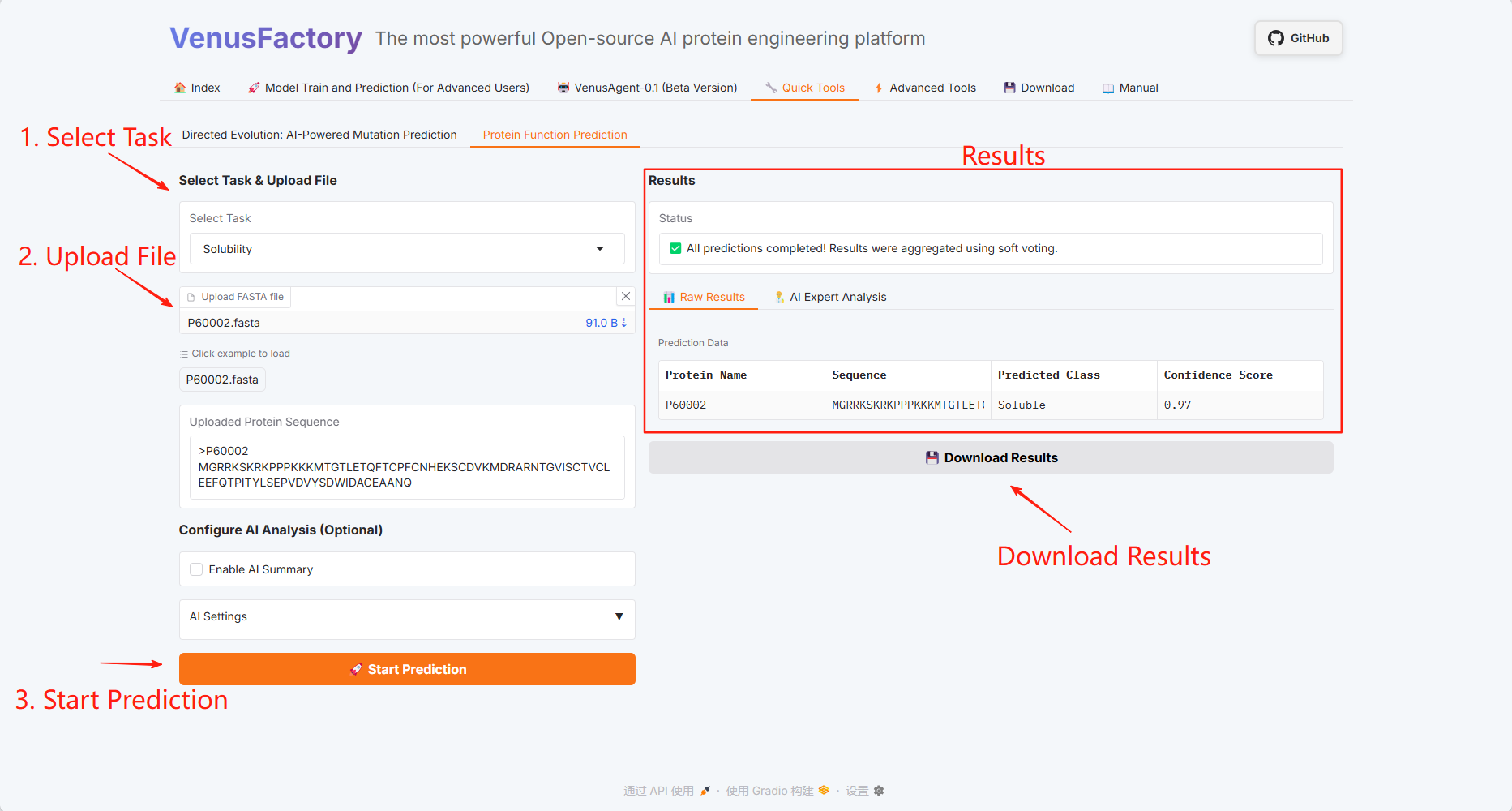
Click the "Quick Tools" module, which includes two functions: Directed Evolution: AI-Powered Mutation Prediction and Protein Function Prediction.
Directed Evolution: AI-Powered Mutation Prediction
Protein Function Prediction
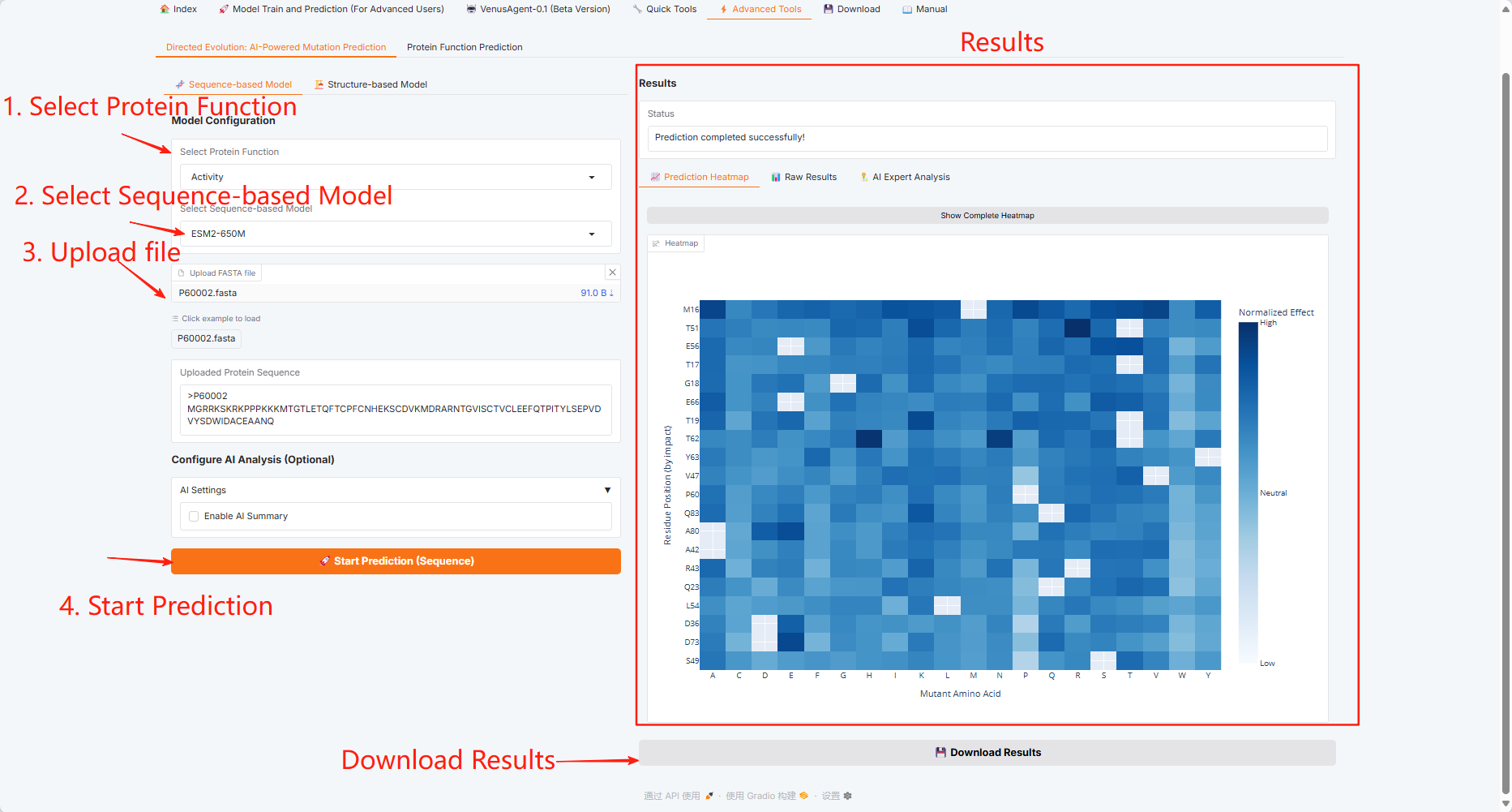
3.6 Advanced Tools
Click the "Advanced Tools" module, which includes two functions: Directed Evolution: AI-Powered Mutation Prediction and Protein Function Prediction.
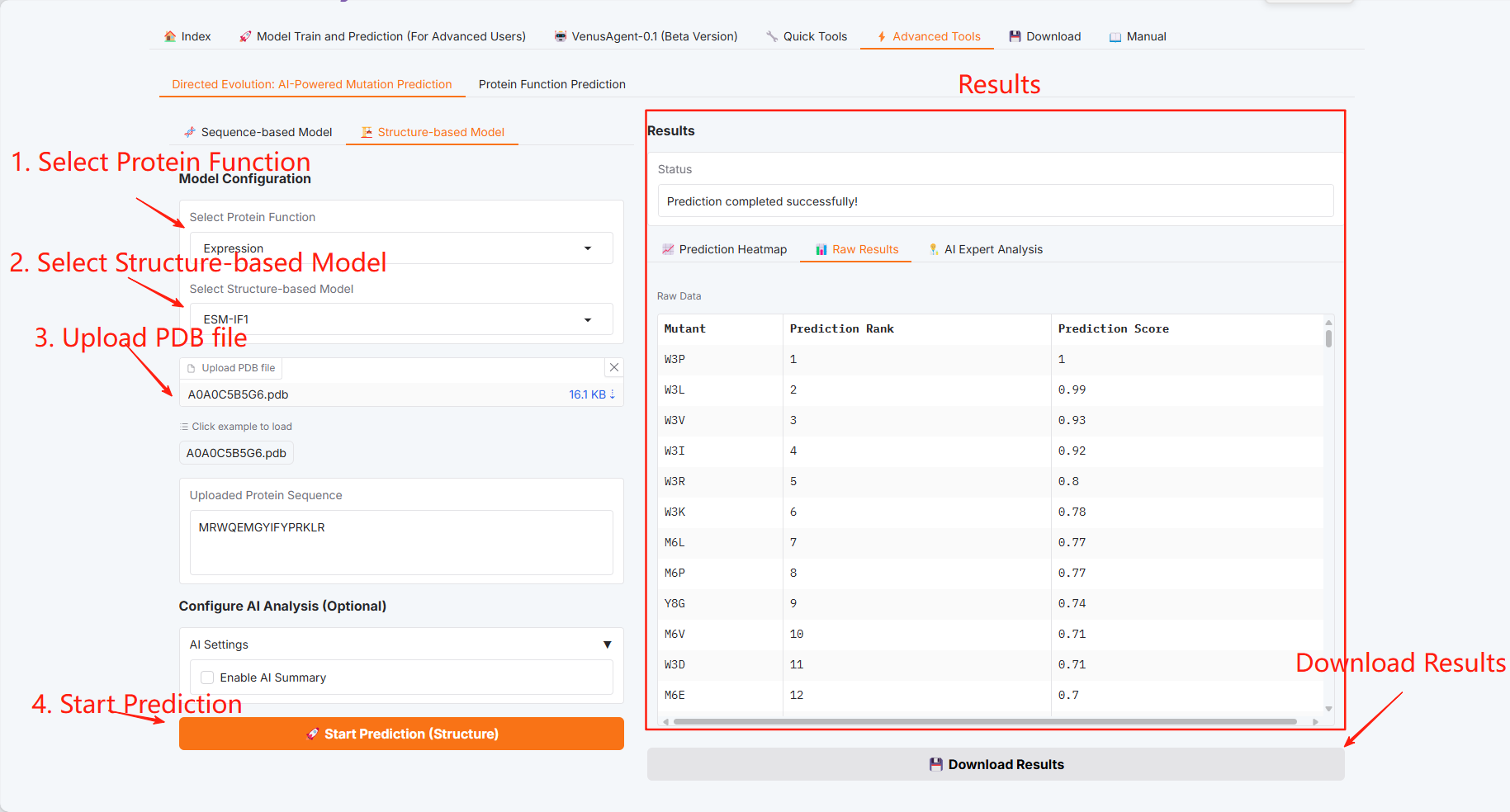
Directed Evolution: AI-Powered Mutation Prediction
Sequence-based Model
Structure-based Model
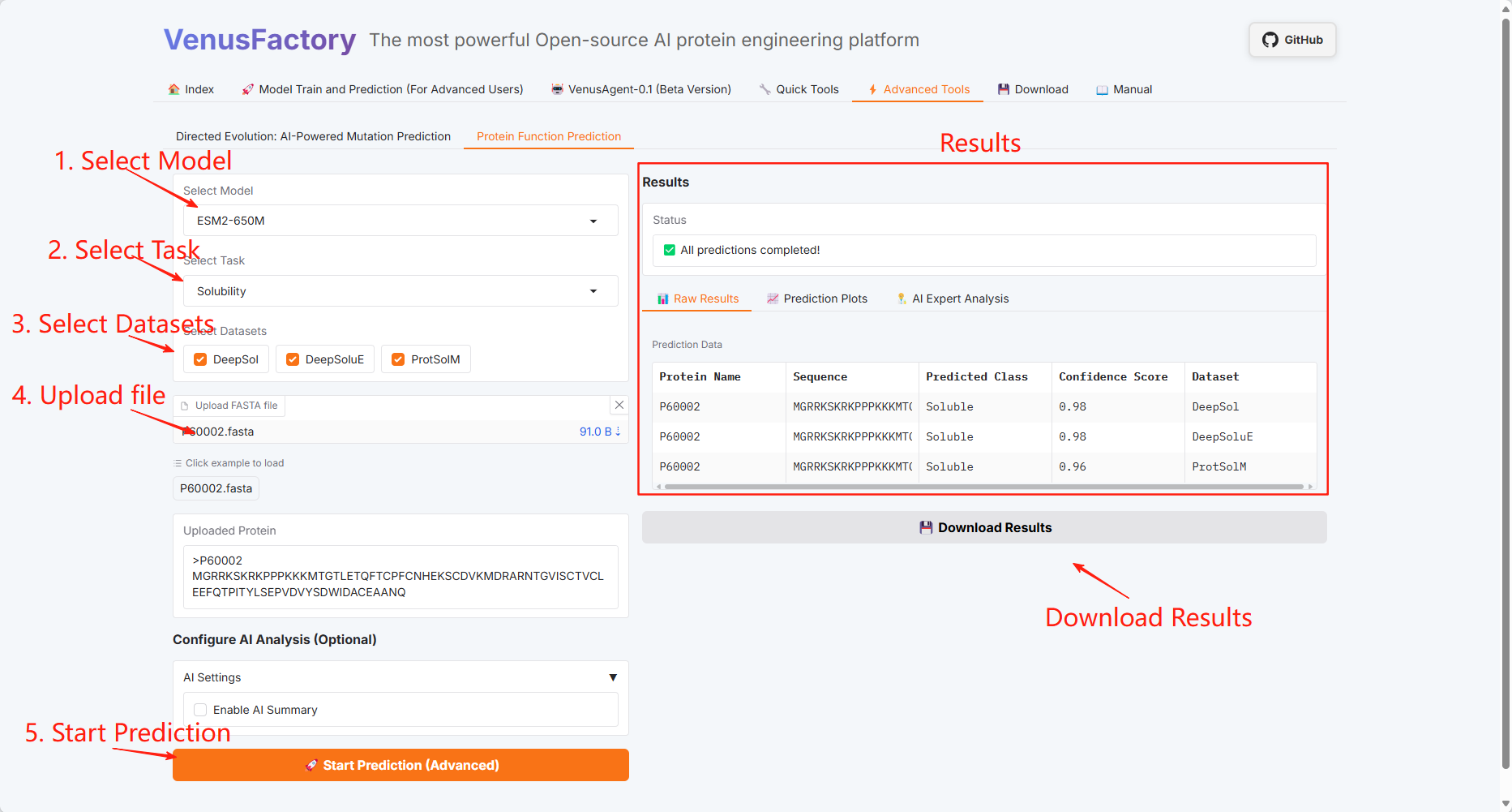
Protein Function Prediction
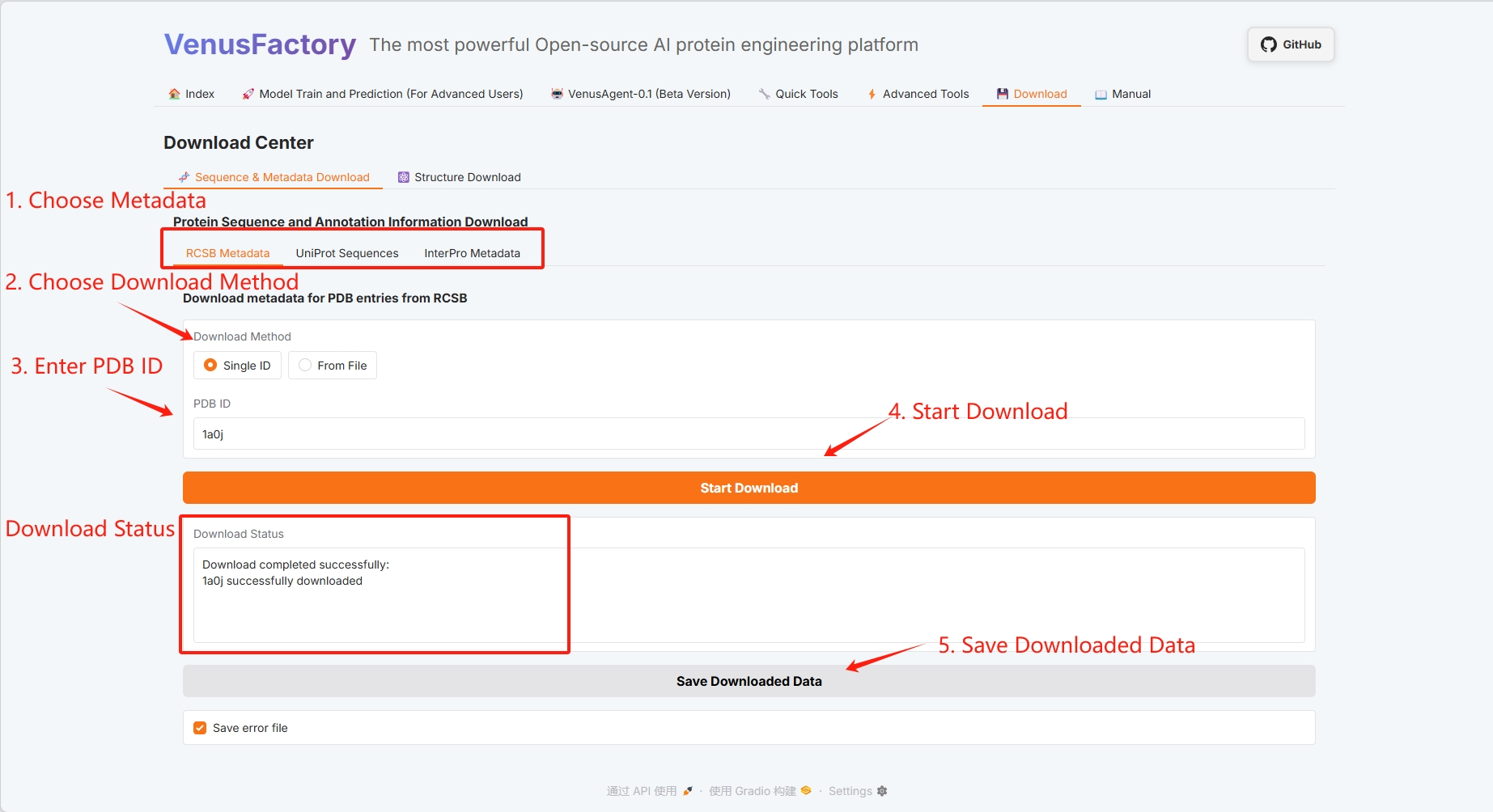
3.7 Download
Click the Download module to download protein data in this interface.
3. Discussion
🖌️ If you see a high-quality project, please leave a message in the background to recommend it! In addition, we have also established an AI4S exchange group. Welcome friends to scan the QR code and remark [AI4S] to join the group to discuss various technical issues and share application results↓

Citation Information
The citation information for this project is as follows:
@inproceedings{tan-etal-2025-venusfactory,
title = "{V}enus{F}actory: An Integrated System for Protein Engineering with Data Retrieval and Language Model Fine-Tuning",
author = "Tan, Yang and Liu, Chen and Gao, Jingyuan and Wu, Banghao and Li, Mingchen and Wang, Ruilin and Zhang, Lingrong and Yu, Huiqun and Fan, Guisheng and Hong, Liang and Zhou, Bingxin",
editor = "Mishra, Pushkar and Muresan, Smaranda and Yu, Tao",
booktitle = "Proceedings of the 63rd Annual Meeting of the Association for Computational Linguistics (Volume 3: System Demonstrations)",
month = jul,
year = "2025",
address = "Vienna, Austria",
publisher = "Association for Computational Linguistics",
url = "https://aclanthology.org/2025.acl-demo.23/",
doi = "10.18653/v1/2025.acl-demo.23",
pages = "230--241",
ISBN = "979-8-89176-253-4",
}Build AI with AI
From idea to launch — accelerate your AI development with free AI co-coding, out-of-the-box environment and best price of GPUs.