Command Palette
Search for a command to run...
MediCLIP Achieves state-of-the-art Results in Anomaly Detection and Localization Using Minimal Medical Imaging data.
In clinical diagnosis and treatment, medical imaging technologies (such as X-rays, CT scans, and ultrasound) are crucial for doctors' diagnosis. After a patient completes an imaging examination, a radiologist or ultrasound specialist typically interprets the images. This professional interpretation identifies abnormalities, locates lesions, and prepares diagnostic reports, providing critical support for clinical decision-making.
With the development of artificial intelligence technology, medical image analysis is ushering in new changes.Currently, machine learning is capable of performing the task of detecting abnormalities in medical images, that is, distinguishing normal images from abnormal images with lesions and identifying the locations of these lesion areas.This type of technology plays a key role in assisting medical decision-making. Some advanced detection models can even perform as well as professional clinicians on specific tasks, reducing decision-making risks while improving the work efficiency of medical staff.
However, existing medical image anomaly detection methods generally rely on large-scale datasets for training. Although this can achieve higher performance, it also significantly increases development costs.
The general-purpose CLIP model performs well in zero-shot transfer, but its direct application to medical image anomaly detection is limited by domain differences and scarce annotations. In 2022, a research team at the University of Illinois at Urbana-Champaign proposed the MedCLIP method. Inspired by visual-text contrastive learning models like CLIP, this approach extends this paradigm to medical image classification and cross-modal retrieval by decoupling image-text contrastive learning and incorporating medical knowledge to eliminate false negatives. The method achieves excellent performance even with significantly reduced data volumes.
While MedCLIP has achieved significant results in alleviating data dependency, it was designed for classification and retrieval tasks and is not well-suited for anomaly detection, particularly lesion localization. Meanwhile, recent research has applied CLIP to zero-shot/small-shot anomaly detection, achieving impressive results. However, these methods often require real anomaly images and pixel-level annotated auxiliary datasets for model training, which are difficult to obtain in the medical field.
To address these issues, a research team from Peking University proposed MediCLIP, an efficient small-sample medical image anomaly detection solution.This method only requires a small amount of normal medical images to achieve leading performance in anomaly detection and localization tasks.It can effectively detect different diseases in various medical image types, demonstrating amazing zero-sample generalization capabilities.
"MediCLIP: Anomaly Detection in Small-Sample Medical Images Using CLIP"Experience the highly effective intelligent medical imaging diagnostics method in the Tutorials section of HyperAI's official website (hyper.ai).
Tutorial Link:
Demo Run
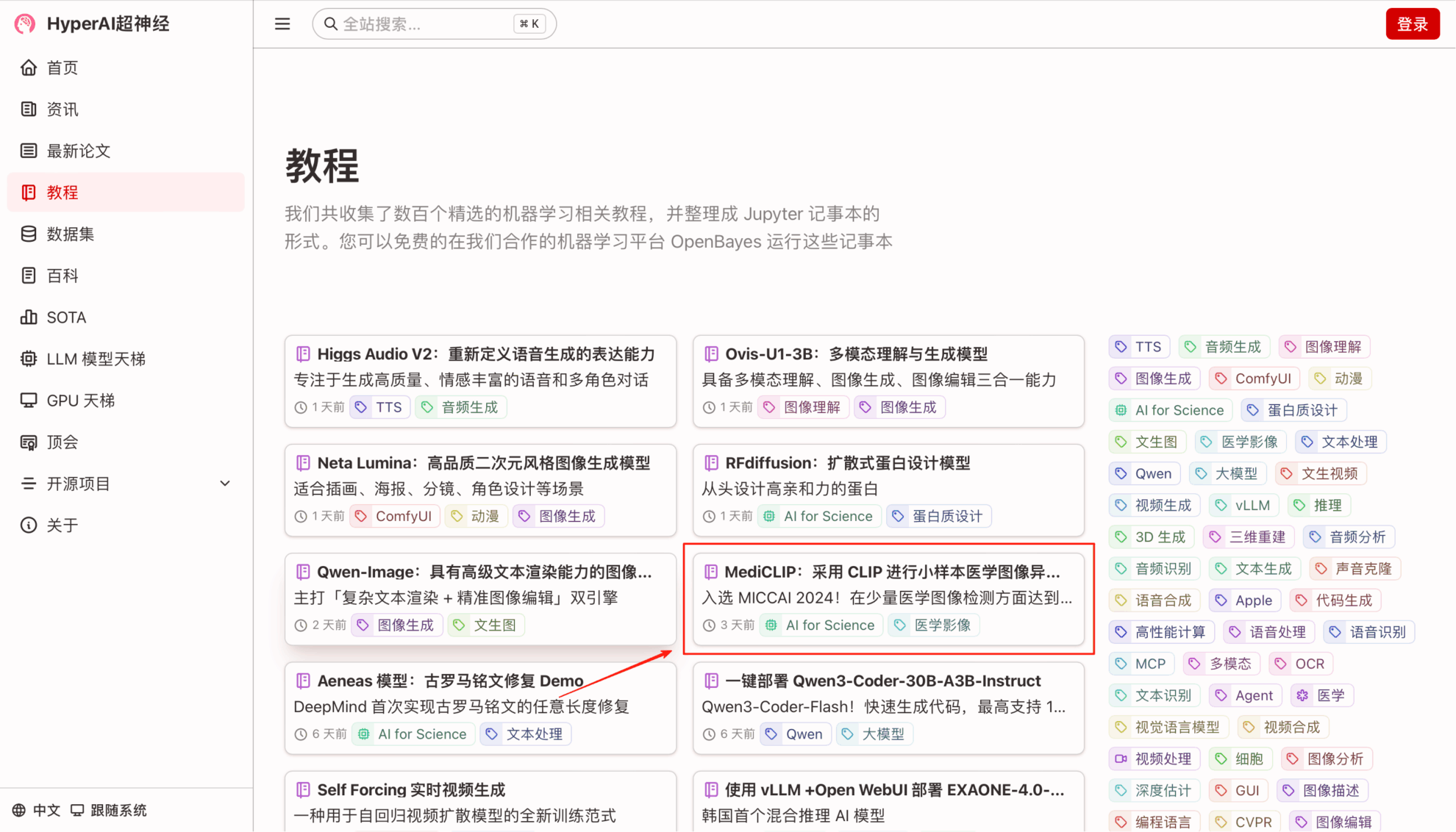
1. On the hyper.ai homepage, select the Tutorials page, choose MediCLIP: Anomaly Detection in Small-Sample Medical Images using CLIP, and click Run this tutorial online.
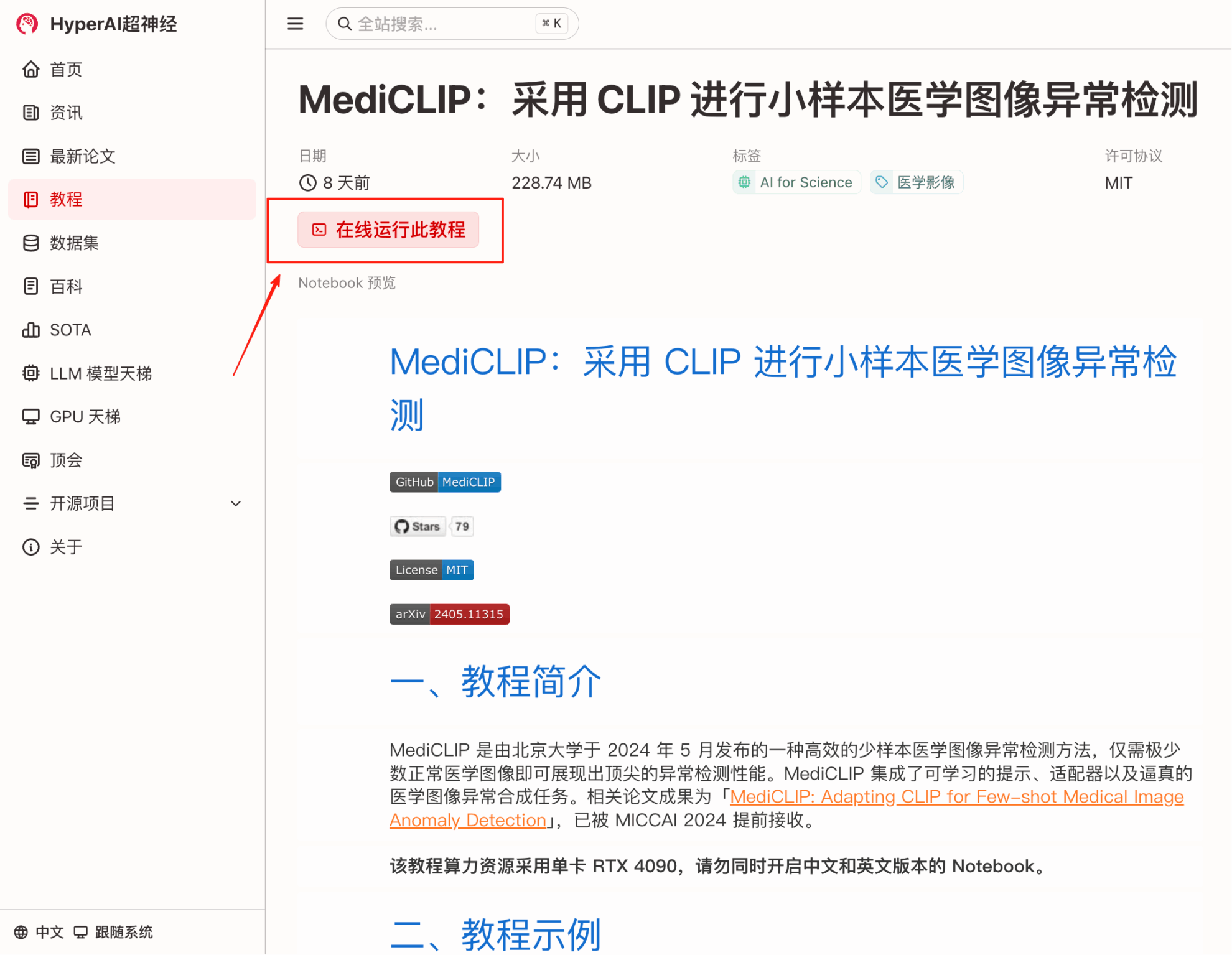
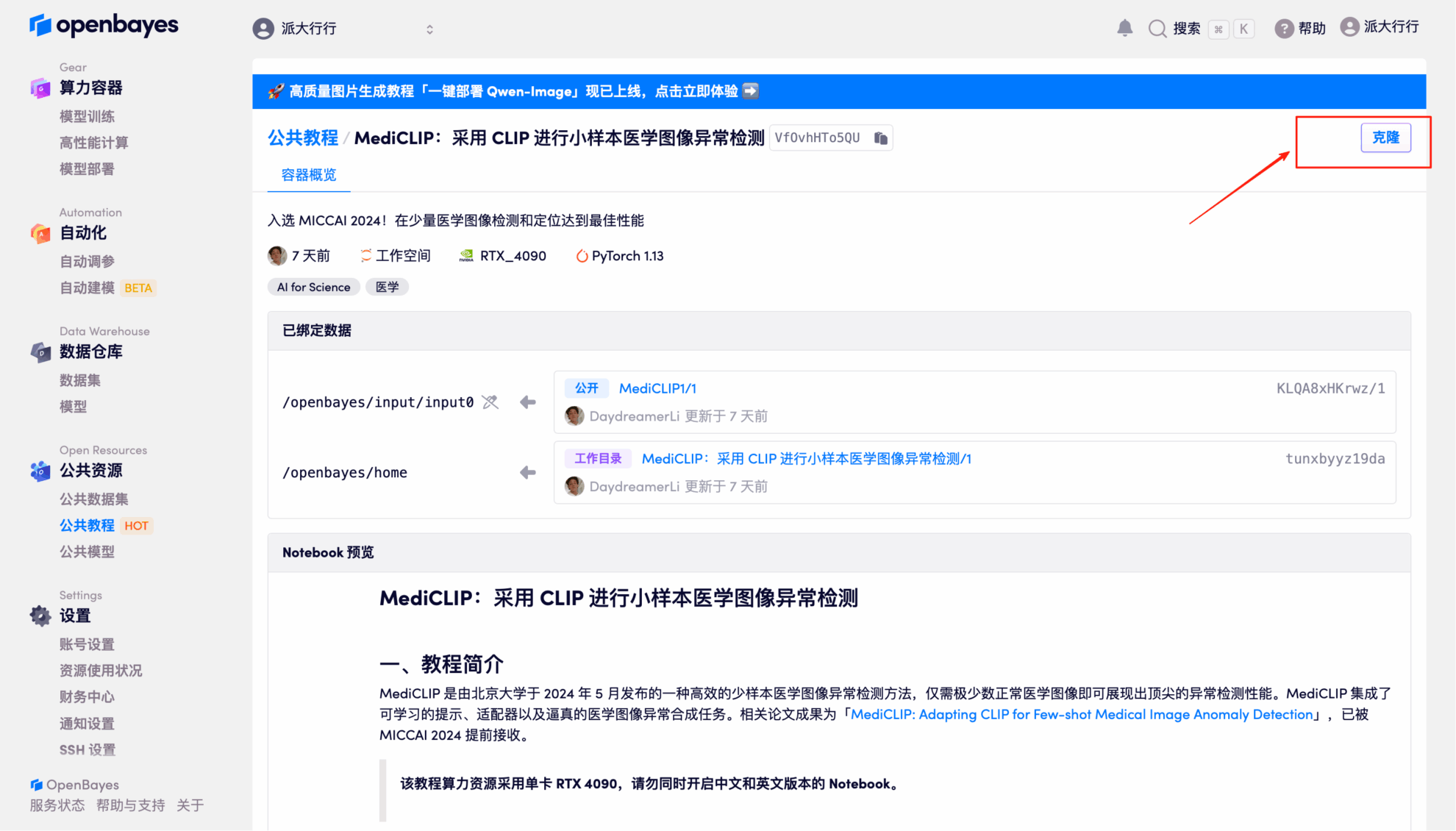
2. After the page jumps, click "Clone" in the upper right corner to clone the tutorial into your own container.
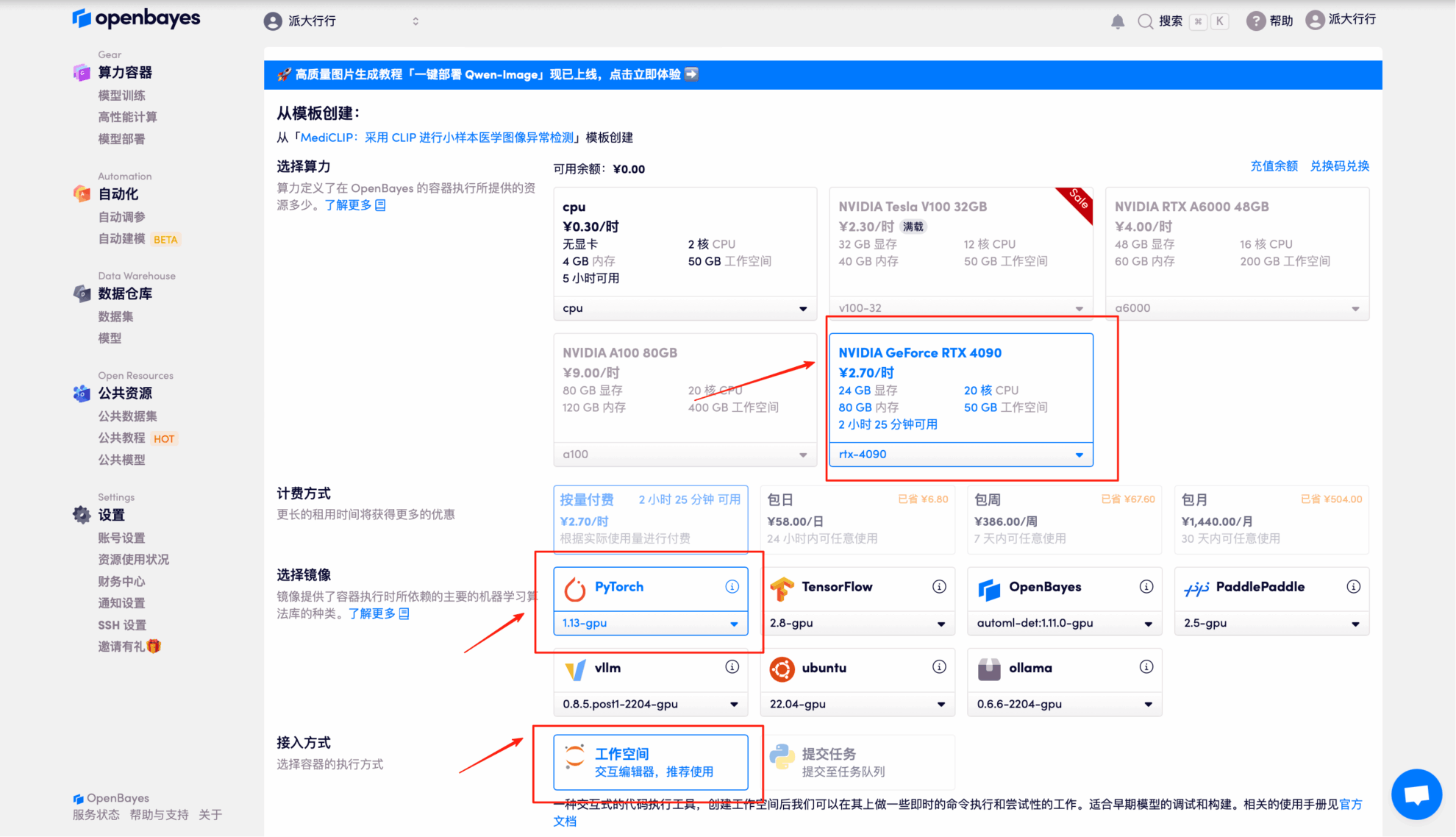
3. Select "NVIDIA GeForce RTX 4090" and "PyTorch" images, and click "Continue". The OpenBayes platform provides 4 billing methods. You can choose "pay as you go" or "daily/weekly/monthly" according to your needs. New users can register using the invitation link below to get 4 hours of RTX 4090 + 5 hours of CPU free time!
HyperAI exclusive invitation link (copy and open in browser):
https://openbayes.com/console/signup?r=Ada0322_NR0n
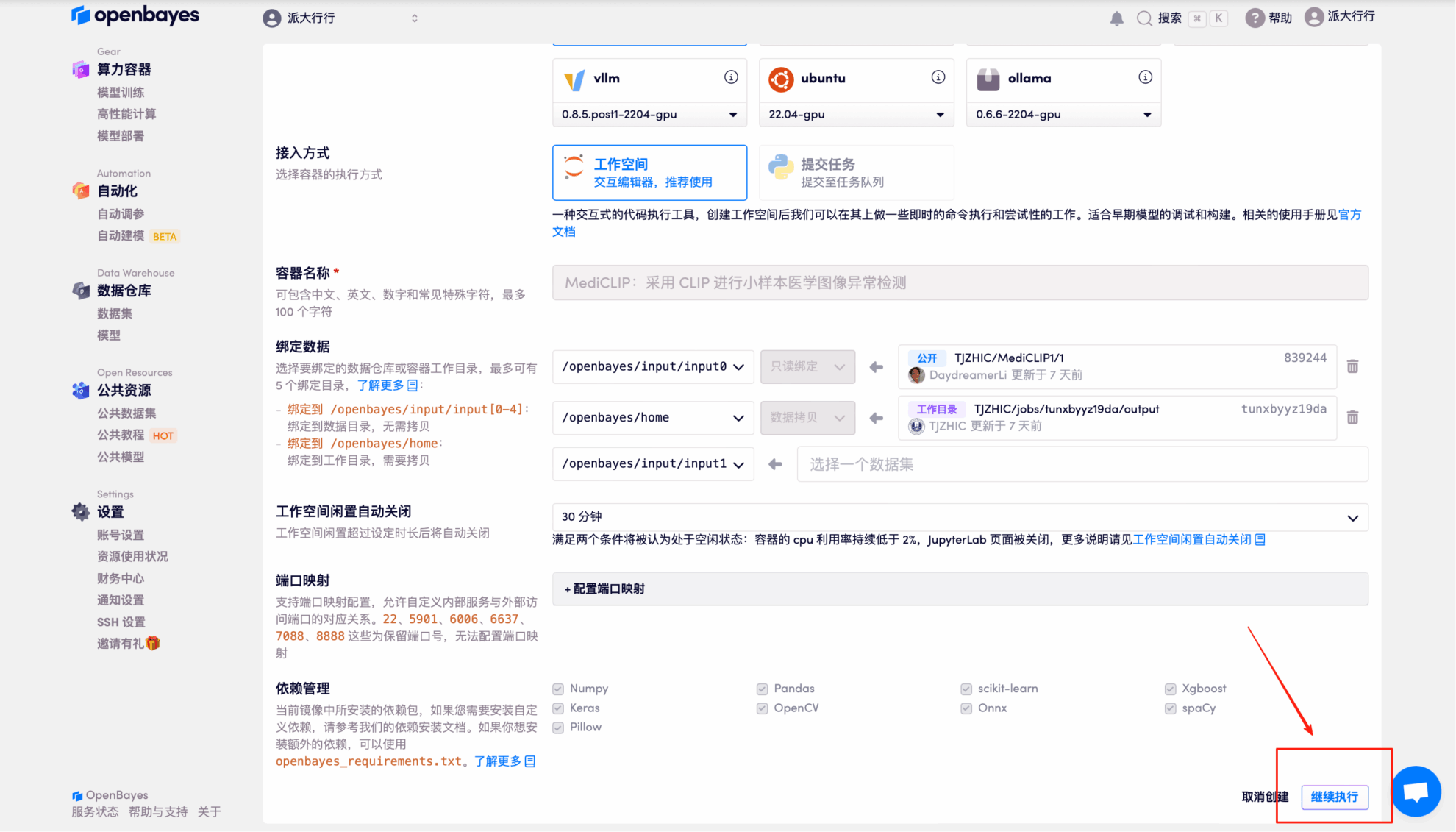
4. Wait for resources to be allocated. The first clone will take about 2 minutes. When the status changes to "Running," click "Open Workspace."
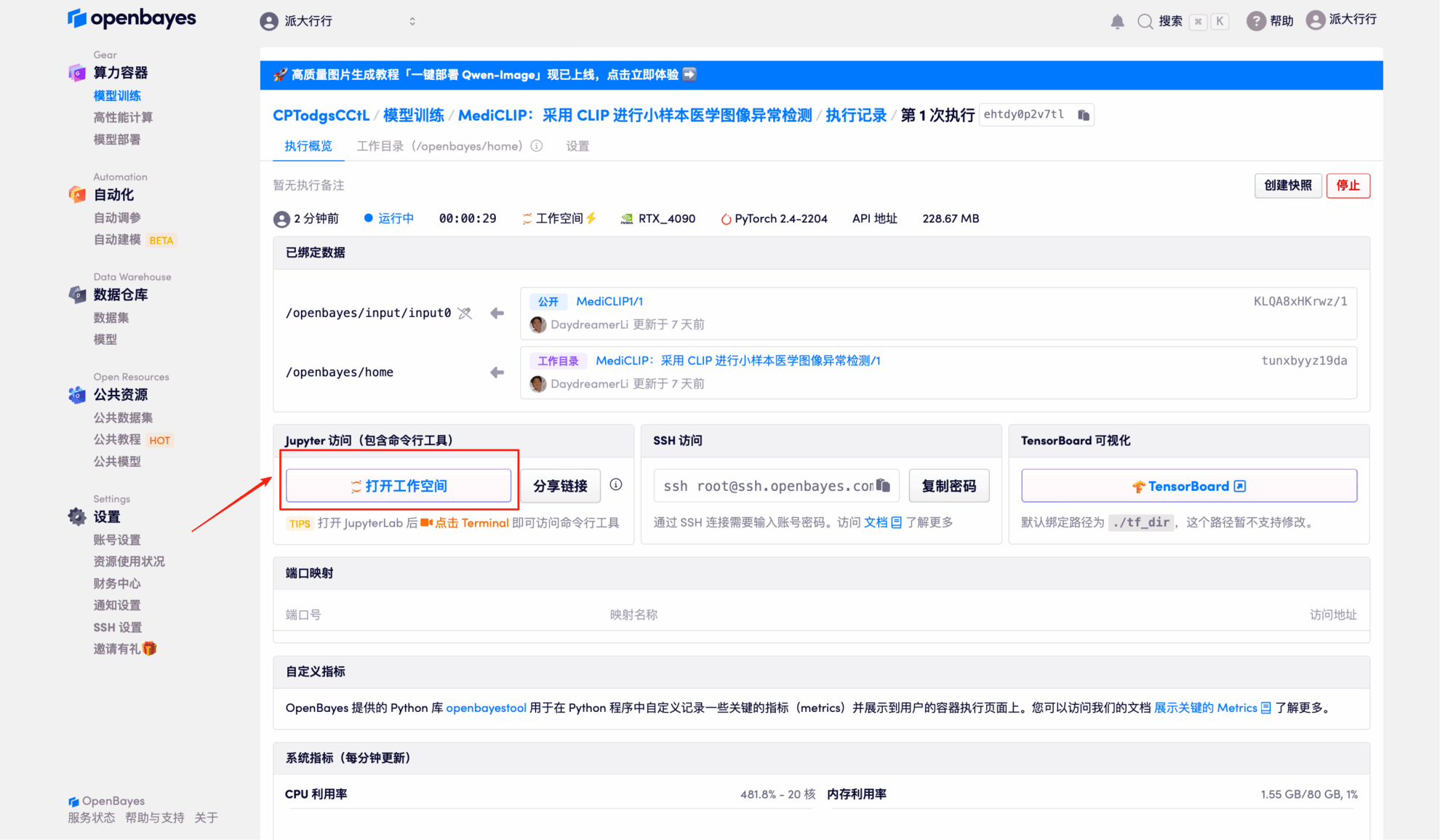
5. Double-click the project name in the left-hand directory to start using it. Note: This tutorial uses a single RTX 4090 GPU. Do not open both the Chinese and English versions of the Notebook at the same time; simply open one. README.ipynb is recommended (the Chinese version is easier to read).
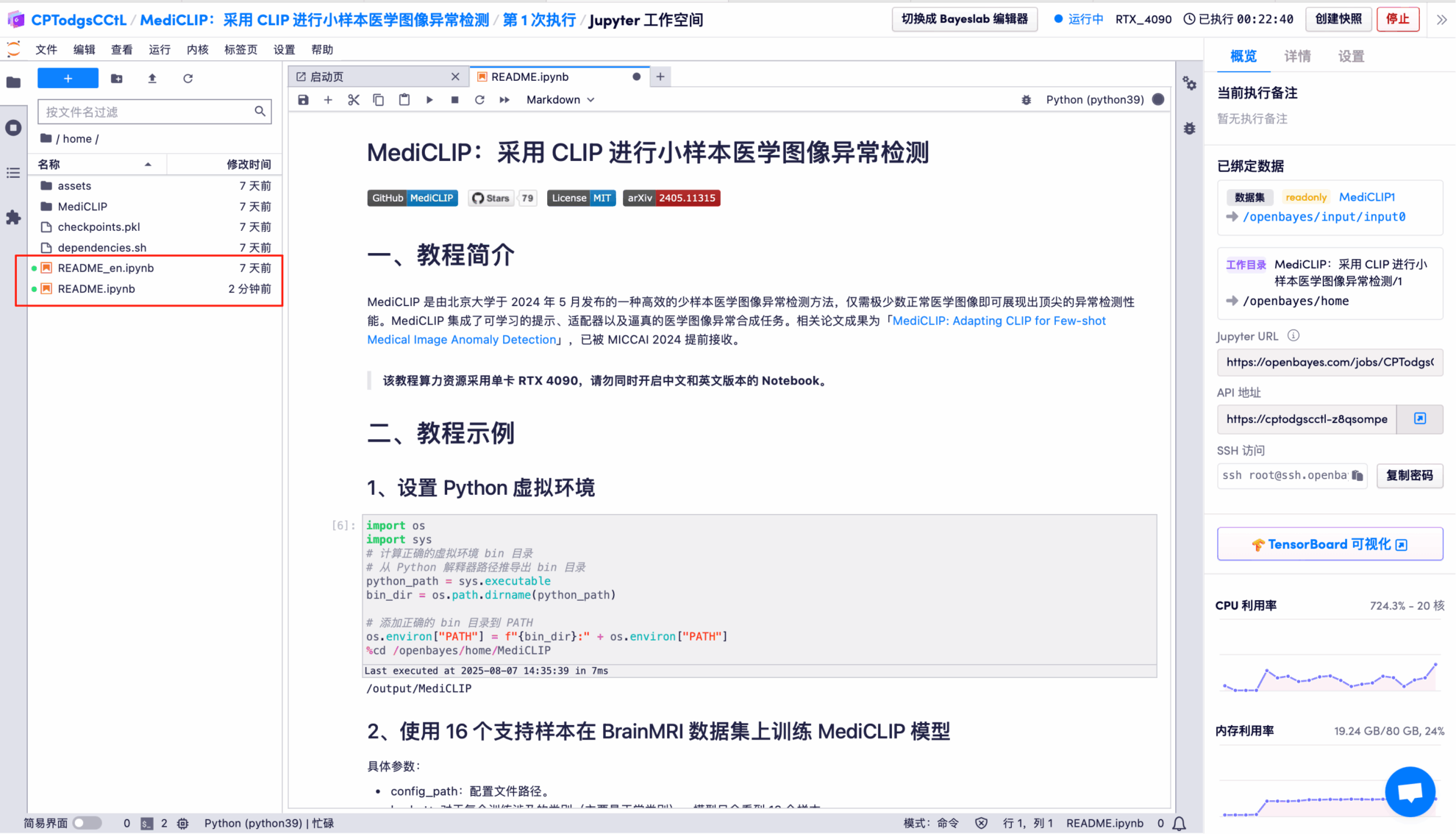
6. Click the "Run button" above: Set up the Python virtual environment.
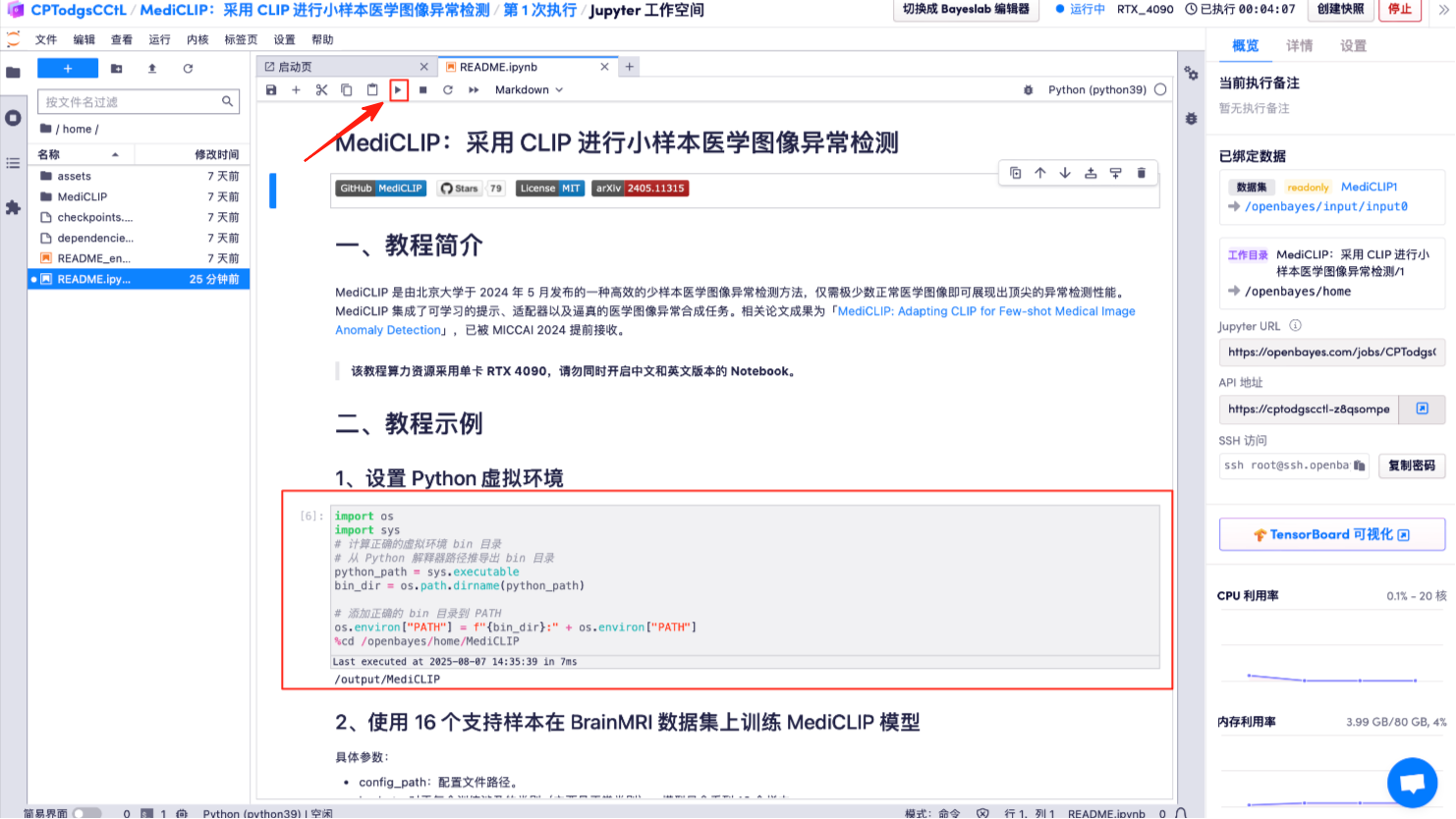
7. MediCLIP model trained on the BrainMRI dataset using 16 support samples.
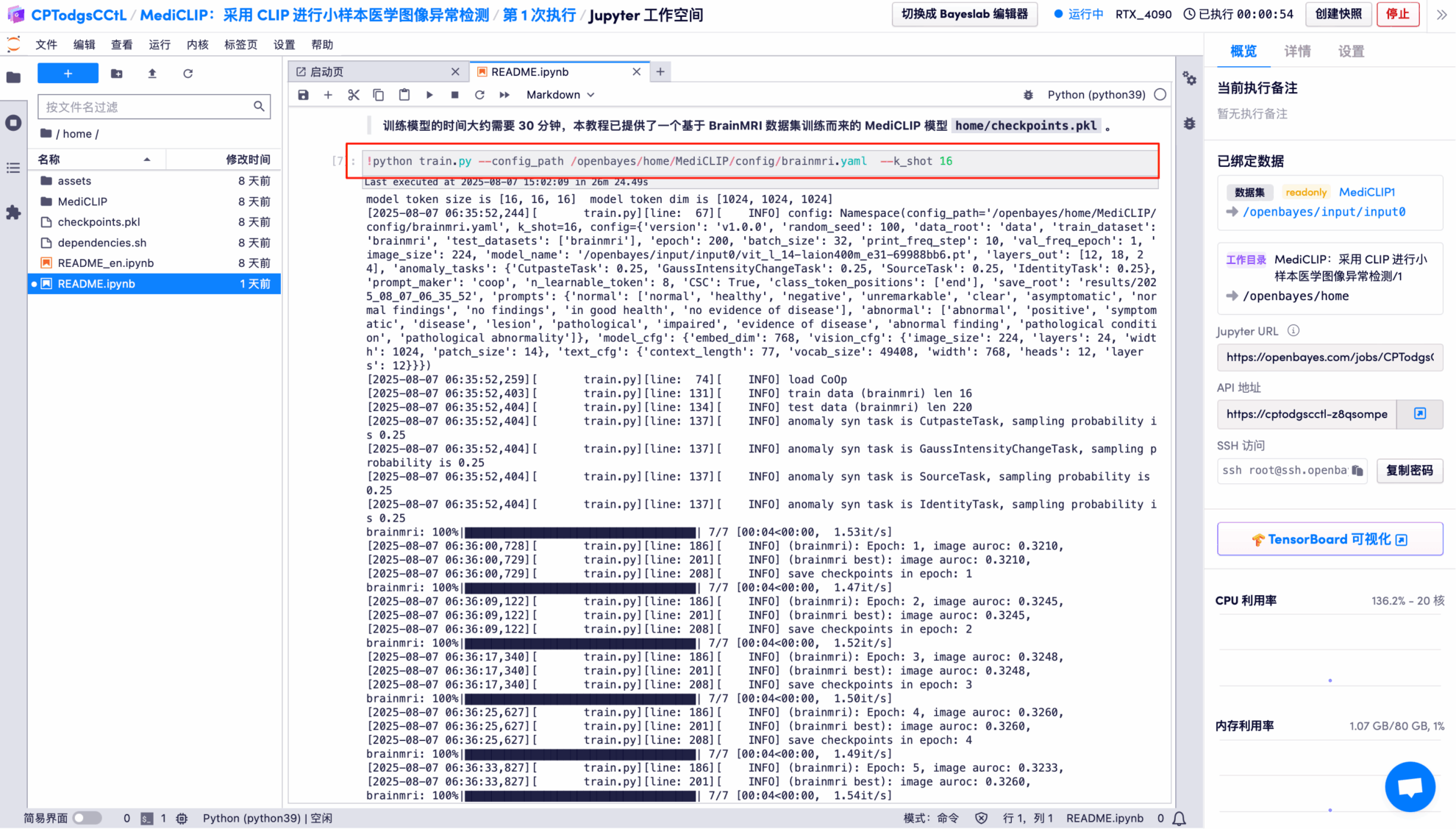
8. Use the trained model to test on the BrainMRI dataset.
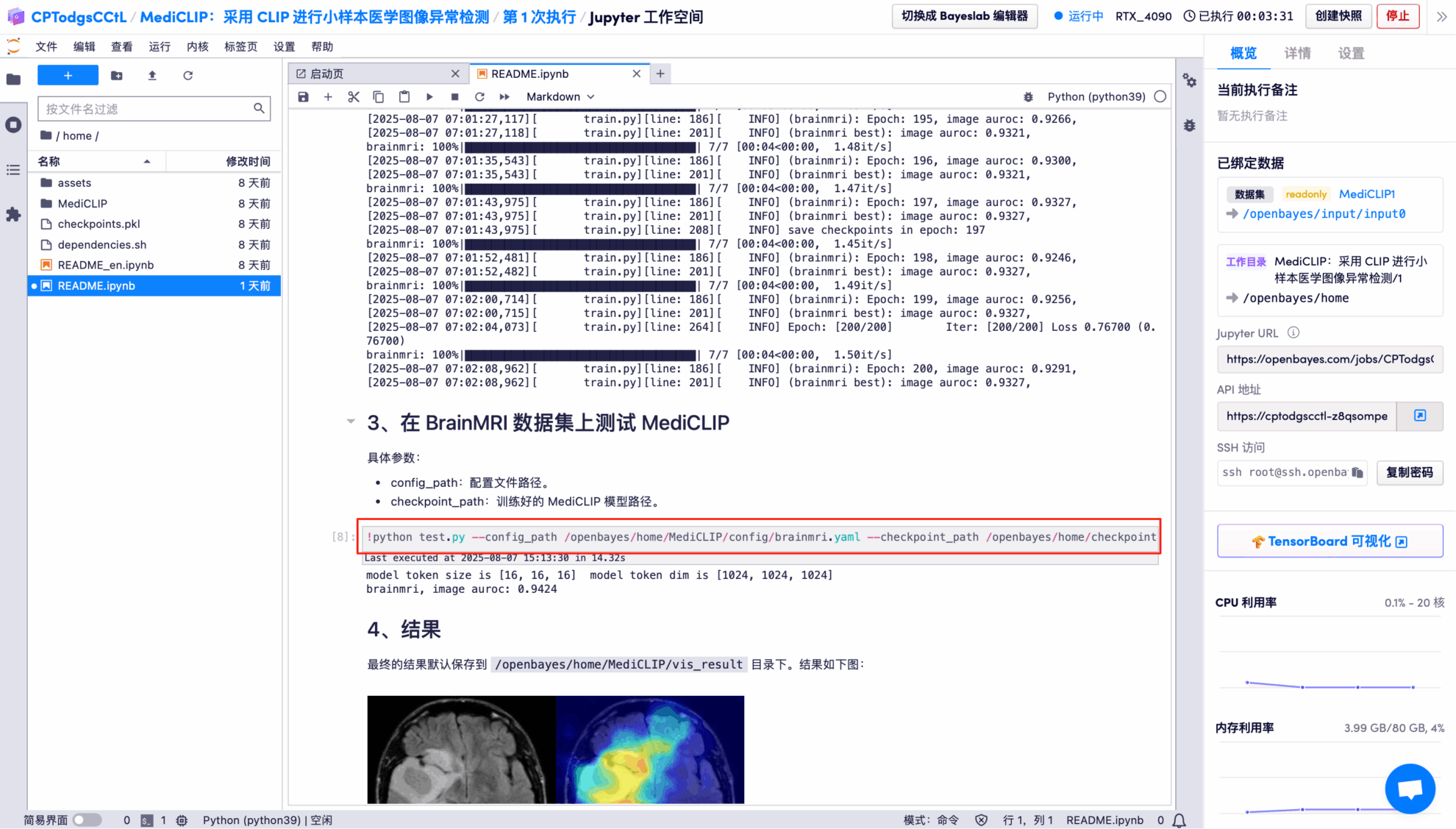
Effect Demonstration
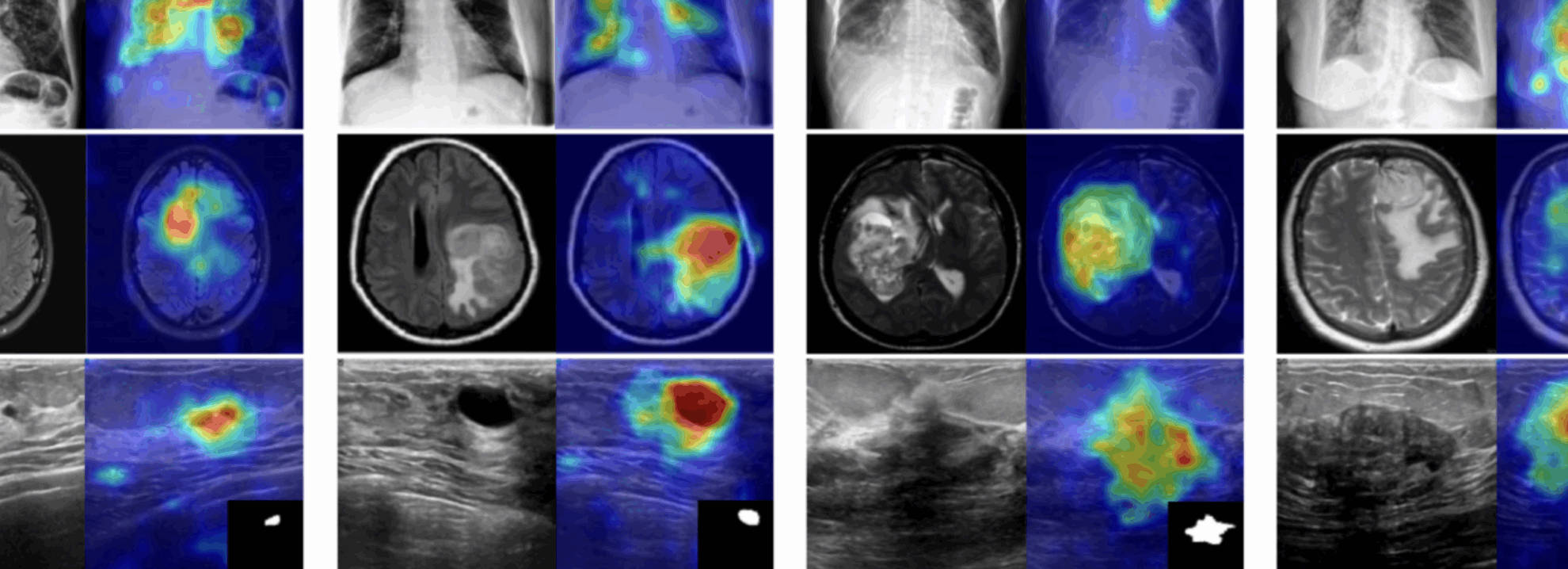
The output results show that the model performs very well in binary classification of images on the BrainMRI test set, with an AUROC of 0.9424 (close to a full score of 1).
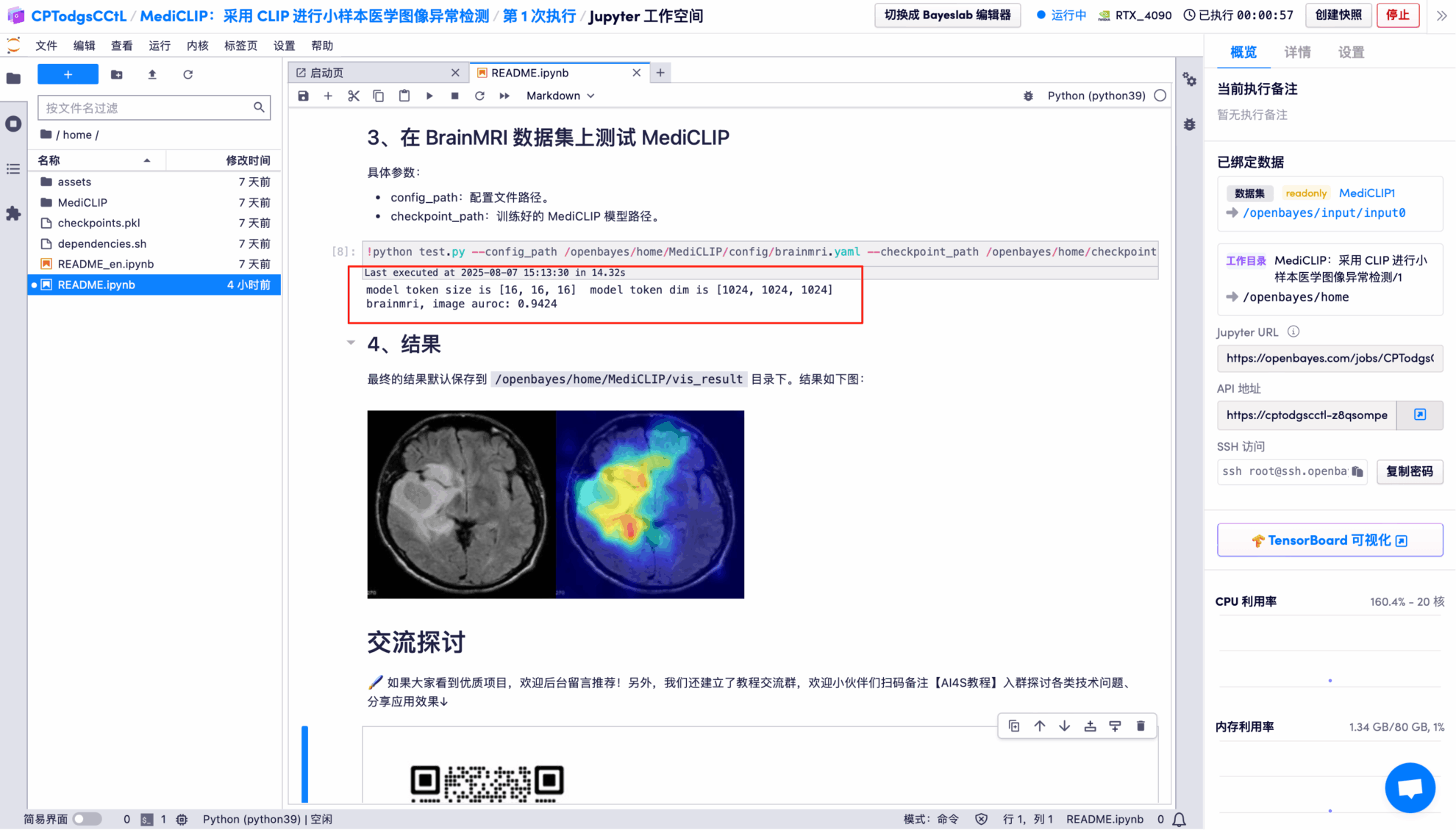
The model's visual detection results for each test image are saved to the openbayes//home/MediCLIP/vis_result directory by default. The specific location is shown in the figure below:
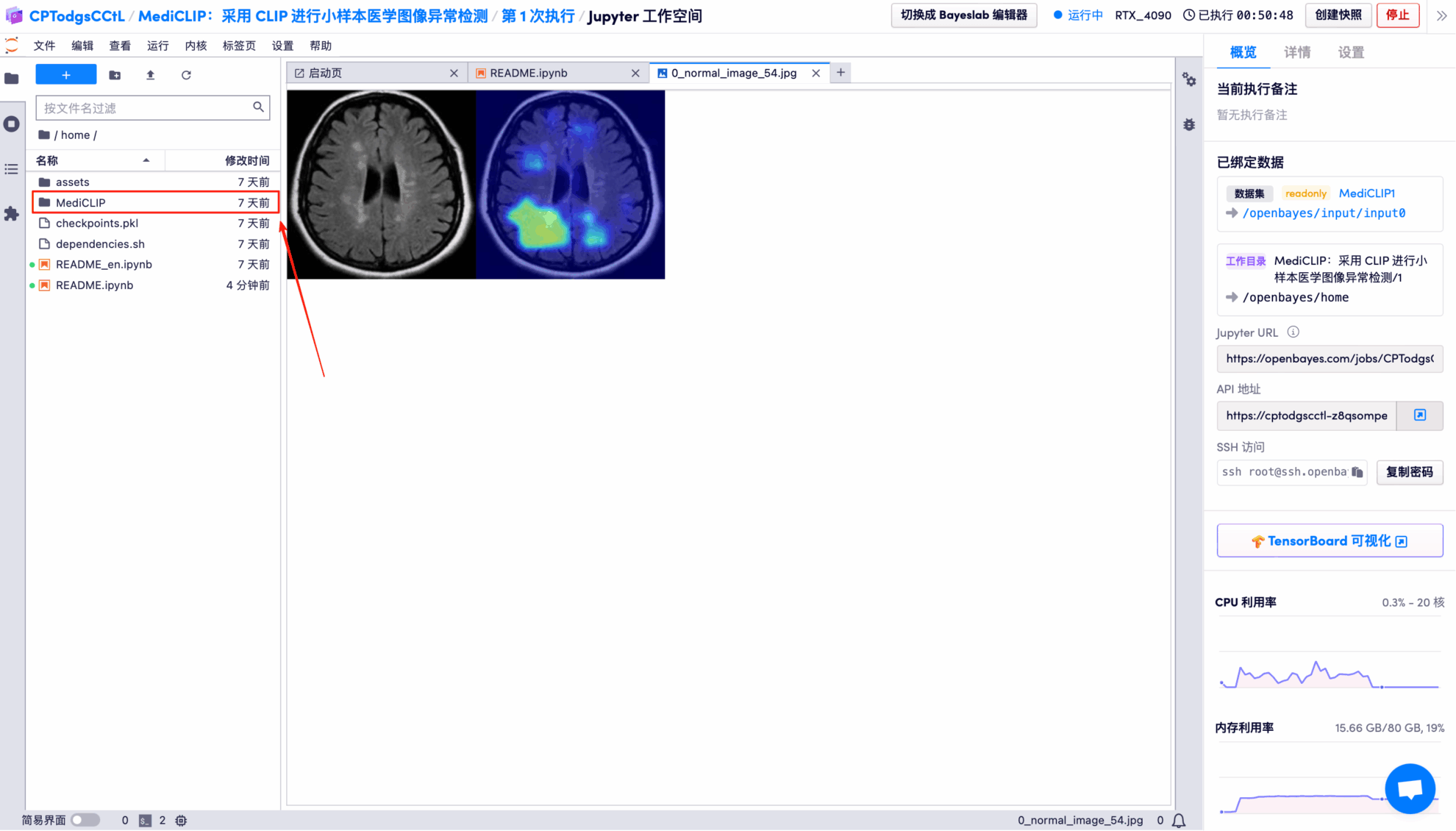
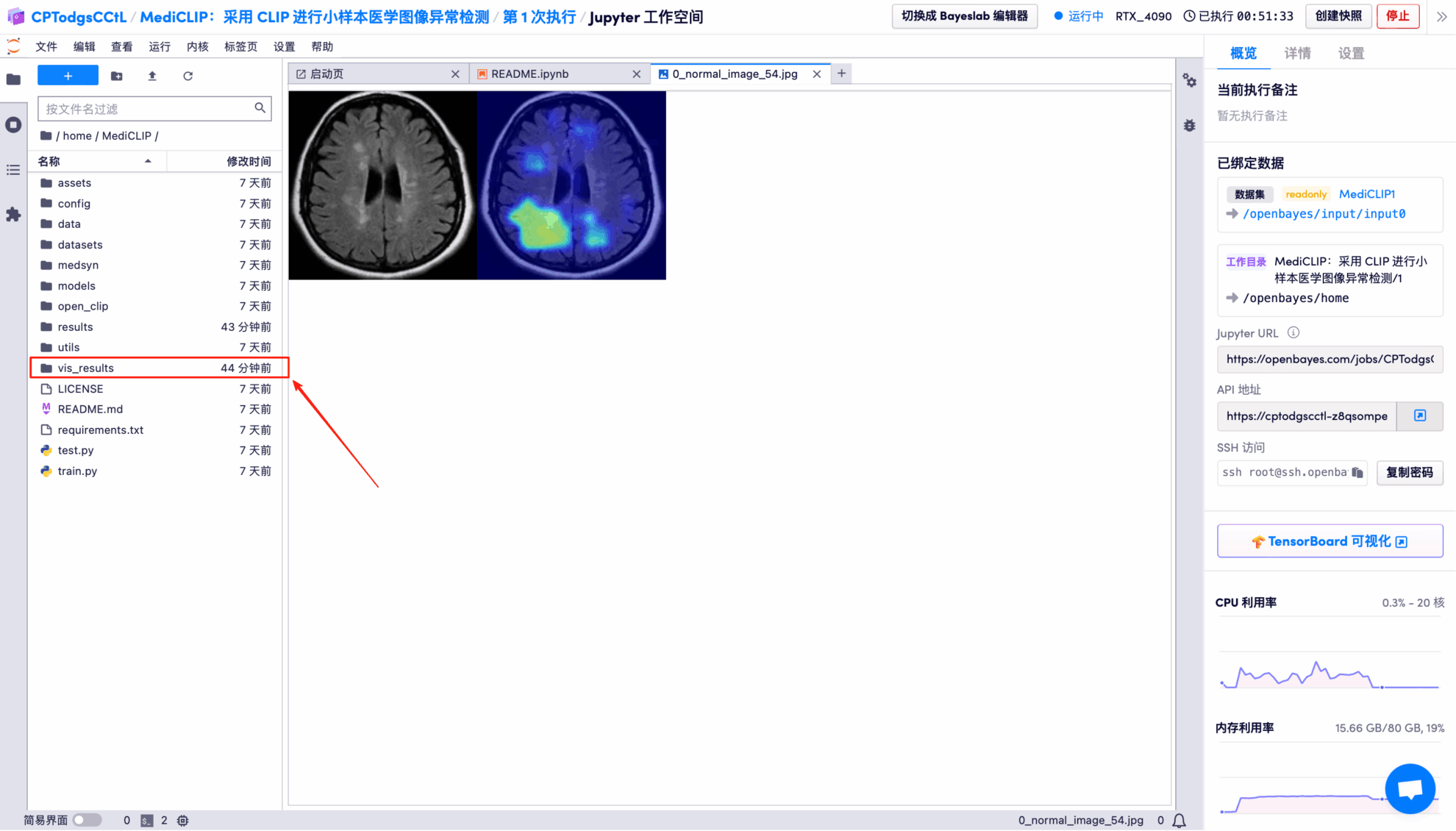
Randomly open a test result picture, which is clearly and intuitively displayed:
* Left: Original brain image
* Right: Highlighted "abnormal area"
The above is the tutorial recommended by HyperAI this time. Everyone is welcome to come and experience it!
Tutorial Link: