Command Palette
Search for a command to run...
The University of Pennsylvania Has Discovered 386 New Antimicrobial Peptides From Animal Venoms and Developed a Deep Learning Model, APEX, to Screen Potential Antibiotic candidates.
The discovery of penicillin in 1928 ushered in a new era in humanity's fight against bacterial infections. These antibiotics significantly enhanced humanity's ability to combat infectious diseases. However, with the widespread and often overuse of antibiotics in fields like medicine and agriculture, bacterial resistance has emerged and is worsening at an alarming rate.
Entering the 21st century, antibiotic resistance has become a major challenge in the field of global public health. According to data from the World Health Organization, approximately 5 million people die each year worldwide directly or indirectly from drug-resistant infections, and the core driver of this crisis is the rapid growth and spread of antibiotic-resistant pathogens. Among them, Gram-negative bacteria, which are listed as "priority pathogens" by the WHO, are particularly troublesome. With their strong ability to evolve drug resistance, they continue to have the upper hand in the "antibacterial war" with humans. What is more serious is that in the past few decades,The research and development of new antibiotics has stagnated: the high R&D costs and long clinical trial cycles have deterred many pharmaceutical companies from entering this field. The number of new antibiotics has sharply decreased, far behind the evolution of drug-resistant bacteria.
Faced with this dilemma, scientists have begun to turn their attention to a potential "treasure trove" in nature - animal venom. After millions of years of natural evolution, animal venom has bred extremely rich molecular diversity, including bioactive peptides and proteins.It can not only interact with a wide range of biological targets, but also exhibit significant antibacterial activity.In fact, the application of venom in biomedical science is already a well-established success story. Ziconotide (trade name Prialt®), extracted from cone snail venom, has become an important analgesic for treating chronic pain; captopril, developed from snake venom, is a commonly used antihypertensive drug in clinical practice. These examples provide important insights into the exploration of venom in the antimicrobial field.
In this context,A research team from the University of Pennsylvania in the United States screened antimicrobial candidate peptides from venom proteins worldwide in a high-throughput manner and discovered 386 antimicrobial peptides with novel structures from animal venoms worldwide.Among them, the experimentally verified molecule 91.4% exhibited strong antibacterial activity, providing a breakthrough idea for the discovery of a new generation of antibiotics.
The relevant research results were published in Nature Communications under the title "Computational exploration of global venoms for antimicrobial discovery with Venomics artificial intelligence."
Research highlights:
* This study constructed a global venom database and generated a candidate library of encrypted peptides, which included a total of 16,123 venom proteins and generated more than 40 million venom encrypted peptides;
* This study used the deep learning model APEX to predict the minimum inhibitory concentration of each peptide against 34 bacterial species, ultimately screening out 386 candidate peptides with antibacterial potential and low sequence similarity to known AMPs;
* The research team synthesized and tested 58 peptides from 386 VEPs, of which 91.4% (53) had significant inhibitory effects on at least one pathogen.

Paper address:
https://www.nature.com/articles/s41467-025-60051-6[*](https://arxiv.org/pdf/2507.09466*)
Follow the official account and reply "APEX" to get the complete PDF
Database: Collection of 16,000 venom proteins
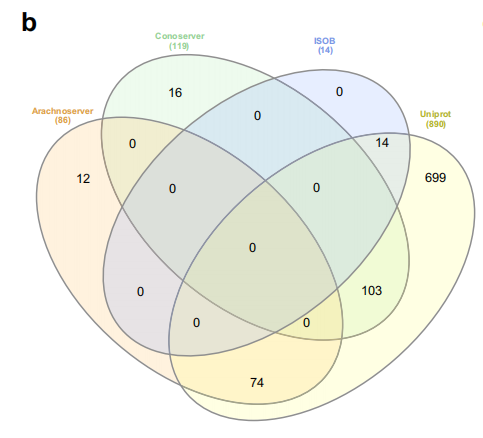
To identify new antimicrobial candidates, the study collected venom protein sequences from four databases: ConoServer, ArachnoServer, ISOB, and VenomZone, yielding a total of 16,123 venom proteins. ConoServer focuses on conopeptides, ArachnoServer includes spider toxins, ISOB is a database of venoms from native snakes of Bangladesh, and VenomZone, compiled from UniProtKB, covers proteins from six taxa: snakes, scorpions, spiders, cone snails, sea anemones, and insects, represented in this study by UniProt.
Each of these datasets has unique characteristics and uses. As shown in the figure below, an analysis of species overlap across different databases reveals that UniProt contains the largest number of unique species, reaching 699, indicating a broader protein diversity. ConoServer and ArachnoServer, on the other hand, contain relatively small subsets of unique species, while ISOB has no unique species at all.The existence of this diversity allows researchers to explore antimicrobial peptides in venom from multiple angles, integrate protein information from different sources, and increase the possibility of discovering new antimicrobial peptides.
As shown in the figure below, researchers processed these venom proteins to generate over 40 million venom encrypted peptides (VEPs). The APEX deep learning model was then used to predict the antimicrobial activity of these peptides, estimating the minimum inhibitory concentration (MIC) value for each peptide against bacterial strains, with the median MIC used as the measure of antimicrobial activity. Through further screening, the researchers ultimately identified 7,379 VEPs with a median MIC ≤ 32 μmol L⁻¹. After filtering for sequence similarity with known antimicrobial peptides, 386 candidates were identified. These candidates differ structurally and functionally from known antimicrobial peptides, providing valuable targets for subsequent experimental validation.
APEX model prediction: a process for integrating multi-source venom proteome data and screening antimicrobial candidate peptides
The APEX model is a deep learning tool that focuses on the potential mining and prediction of antimicrobial peptides.Its core advantage lies in its ability to predict bacterial strain-specific antimicrobial activity. After systematic training, the model can accurately predict the minimum inhibitory concentration (MIC) values of peptides for 34 different bacterial strains—a key parameter for measuring the effectiveness of antimicrobial agents.
The training dataset of the model consists of two parts:Internally accumulated peptide sequence data and public antimicrobial peptide (AMPs) information obtained from the DBAASP database.This diversified data support enables APEX to perform outstandingly in analyzing the complex mapping relationship between peptide sequences and antibacterial activity, laying a solid algorithmic foundation for subsequent screening.
In terms of architectural design, APEX adopts a multi-objective task model framework and a "three-step" strategy to screen antimicrobial peptide candidate molecules from venom proteins.first,The sliding window method was used to construct a peptide library, and fragments with lengths ranging from 8 to 50 amino acids were selected.Secondly,The APEX model was used to predict the minimum inhibitory concentration (MIC) of these peptides against different bacterial strains;at last,Further selection based on sequence similarity resulted in a group of venom encrypted peptides (VEPs) with novel structures and excellent activity.
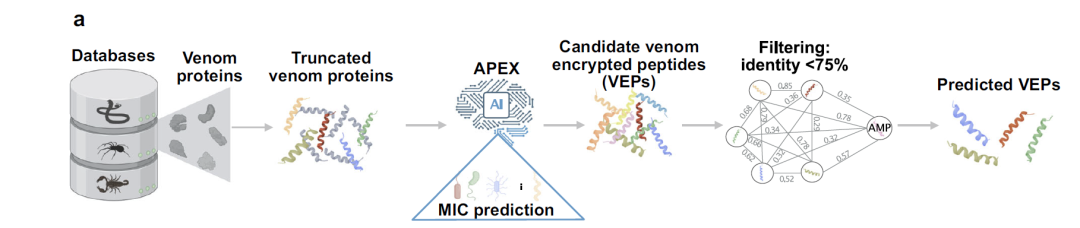
This design enables the precise identification of candidate molecules with potential antimicrobial activity when processing massive amounts of venom encrypted peptides (VEPs). Compared to traditional experimental screening methods, APEX significantly improves the efficiency of antimicrobial peptide discovery while reducing reliance on resource-intensive biochemical assays, providing researchers with a faster and more economical screening path.
In practical applications, using the APEX model requires certain technical preparation. Researchers need to set up a suitable runtime environment, including installing Python 3.9, a specific version of PyTorch, and a series of dependent libraries such as numpy, scipy, and matplotlib. Once the environment is set up, the candidate peptide sequences are saved in a text file. By running the specified command, the model generates a CSV file containing the MIC prediction results. By analyzing these predictions, researchers can quickly identify peptides with high antimicrobial potential for further experimental verification.
Verification of the antibacterial activity of VEPs and analysis of their mechanism of action
To verify the antibacterial activity of venom encrypted peptides (VEPs), the research team conducted a series of experiments.In the antimicrobial activity assay, 58 VEPs were tested against a variety of pathogens.The results are shown in the figure below, 53 of them (accounting for 91.4%) showed activity against at least one pathogen.All VEPs from the ArachnoServer database showed antibacterial activity, which strongly confirmed the strong antibacterial potential of this type of peptide.

However, some VEPs from the UniProt database had relatively limited efficacy. For example, UniprotKB-2 did not show any antibacterial activity, and UniprotKB-6 and UniprotKB-11 only had inhibitory effects on Enterococcus faecalis. Further analysis revealed that these low-activity peptides generally had low hydrophobicity and net charge - a finding that suggests thatHydrophobicity and net charge play key roles in promoting the interaction between peptides and bacterial membranes, and their levels may directly affect the antibacterial efficacy of VEPs.
In the secondary structure study, the team observed through circular dichroism (CD) analysis that the structural changes of VEPs are significantly dependent on the environment. In aqueous solution, VEPs mainly exist in a disordered structure; however, in a simulated bacterial membrane environment (such as SDS micelle system) or a helical induction medium (such as trifluoroethanol/water mixed solution), they undergo a significant conformational transition, folding from a disordered structure to an α-helix. This structural feature is highly consistent with the behavior of typical antimicrobial peptides.This suggests that VEPs may be specially adapted to biofilm-related functions, which also provides important clues to their antibacterial mechanism.
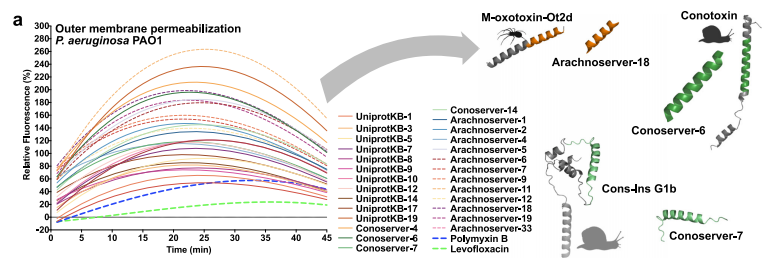
In terms of mechanism of action research, the team evaluated the effects of VEPs on bacterial outer membranes and cytoplasmic membranes through fluorescence assays. The results are shown in the figure below. 23 VEPs can effectively penetrate the bacterial outer membrane, among which Arachnoserver-18, ConoServer-6, and ConoServer-7 have particularly outstanding permeability activities. In the cytoplasmic membrane depolarization experiment, the depolarization effect of most tested VEPs was better than that of the control group, but slightly weaker than that of some known peptide families. Overall,The antibacterial effect of VEPs is mainly achieved by inducing cytoplasmic membrane depolarization. This mechanism is consistent with the action mode of many known antimicrobial peptides and encryption peptides, further supporting its rationality as a new antibacterial agent.
Academia and industry collaborate to explore and develop new low-resistance antimicrobial drugs
In order to develop new antimicrobial drugs that are less likely to induce drug resistance, collaborative exploration between academia and the business community, from basic research to industrial transformation, is constantly unlocking the application potential of venom antimicrobial peptides.
In academia, research teams from various countries have conducted in-depth research on the discovery, design and mechanism analysis of venom antimicrobial peptides. The Mu Yuguang team at Nanyang Technological University in Singapore has addressed the problem that traditional models are limited by the amount of data and feature dimensions in predicting protein interactions.Constructed the largest structure-based protein interaction dataset to date,They also developed the ProAffinity-GNN deep learning framework. This model innovatively integrates protein language models with graph neural networks, deeply coupling sequence information with spatial structural features. Not only does it surpass existing methods in the accuracy of protein-protein affinity prediction, its powerful generalization capabilities also provide a technical paradigm for the efficient prediction of interactions between venom peptides and bacterial targets. Furthermore, the analysis of the mechanism of action provides theoretical support for the optimized design of antimicrobial peptides.
The team from Qilu Medical College of Shandong University used the diffusion model in the field of artificial intelligence to simulate the "random perturbation-directed screening" process of peptide evolution in nature.40 novel peptide sequences were successfully generated, 25 of which showed clear antibacterial or antifungal activity.
In the business world, Israel's Bountica Company focuses on the pain points in the field of food preservation, and uses microbial fermentation technology to mass-produce antifungal peptides derived from venom. Its VenomShield series of products,Only one part per million is needed to inhibit the growth of mold and yeast in bread and juice, extending the shelf life to more than twice that of traditional preservatives.
China Shanghai Hi-Tech Bioengineering Co., Ltd. cooperates with Fudan University,Together, we have built three relatively complete antimicrobial protein databases in the world.These databases not only provide efficient tools for the research and development of new biological antimicrobial enzymes and antimicrobial peptides, but also accelerate the research of new substances against drug-resistant microorganisms through systematic data mining.
This demonstrates the tremendous vitality of the research and translation of venom antimicrobial peptides, spanning both basic science and industrial application. As deep learning models increasingly accurately analyze sequence-function relationships, and synthetic biology techniques continuously optimize peptide stability and expression efficiency, these molecules, inspired by nature's "survival wisdom," are poised to become a key force in the fight against antibiotic resistance and provide new safeguards for global public health.
Reference articles:
1. https://mp.weixin.qq.com/s/lwYWWIe9-Az22jlhiZ747A
2. https://mp.weixin.qq.com/s/sGHRf-ebaSRiqGiUxMMueA
3. https://mp.weixin.qq.com/s/0mJltGuaKTUwYDGK4wD9UA
4. https://mp.weixin.qq.com/s/R6Y_-38saZwqSSow0LNhzw