Command Palette
Search for a command to run...
Based on 8 Million Real Data, the Cornell University Team Used Graph Neural Networks to Accurately Predict the Survival of Lung Cancer Patients and Discovered 3 Deadly Subtypes
Ten years ago, the results of the CheckMate 017 trial shocked the oncology community. The New England Journal of Medicine, The Journal of the American Medical Association, and other journals have reported many times that the survival data of patients with advanced squamous cell lung cancer treated with the PD-1 inhibitor Nivolumab has significantly improved: the median overall survival has increased from 6 months with chemotherapy to 9.2 months, and the 18-month survival rate is twice that of the chemotherapy group. This study marks the beginning of the era of immune checkpoint inhibitors (ICI), but it also exposes the problem of large differences in the response of patients with advanced non-small cell lung cancer (aNSCLC) to immunotherapy:In the trial, some patients' tumors continued to remit for more than 3 years, while others experienced disease progression within a few months. This heterogeneity in treatment response has become a problem in the era of precision medicine.
The complexity of lung cancer stems from its high heterogeneity. Non-small cell lung cancer (NSCLC) accounts for 80%-85% of lung cancer.About 75% patients are diagnosed at the advanced stage, and the 5-year survival rate is only 26.4%.Differential expression of tumor microenvironment biomarkers, different functional states of immune cells, and diverse comorbidities of patients make the pathological situation complicated. Patients receiving ICI treatment benefit from high expression of PD-L1, but also have poor efficacy due to low tumor mutation load, and comorbidities may also affect treatment options and prognosis.
To meet the challenges, diagnosis and treatment plans are transitioning from "one size fits all" to "precise stratification." In this transition process, predictive medicine has gradually emerged. Its core goal is to integrate multi-dimensional data, including electronic health records and omics information, so as to tailor the most appropriate treatment plan for each patient. In recent years, with the continuous accumulation of large-scale biomedical data and the rapid development of machine learning technology, researchers have begun to try to use unsupervised machine learning methods to perform cluster analysis on patient groups with similar characteristics in order to predict treatment responses. Unfortunately, however, traditional methods often have limitations in practical applications.It is difficult to ensure consistency in survival outcomes among patients within the group, which limits the application value of stratified results in clinical practice.
To solve the above problems, Cornell University and Regeneron Pharmaceuticals proposed the Graph Encoded Mixed Survival Model (GEMS).Complex relationships in patients’ electronic health records were encoded through graph neural networks and combined with survival analysis models to identify subphenotypes with consistent characteristics and survival outcomes.The study found that it is superior to traditional methods in predicting overall survival (OS), identifying three sub-phenotypes with different clinical characteristics and survival patterns, opening up a new path for precision medicine for lung cancer.
The relevant research results have been published in Nature Communication under the title "Identification of predictive subphenotypes for clinical outcomes using real world data and machine learning".

Paper address:
https://doi.org/10.1038/s41467-025-59092-8
The open source project "awesome-ai4s" brings together more than 100 AI4S paper interpretations and also provides massive data sets and tools:
https://github.com/hyperai/awesome-ai4s
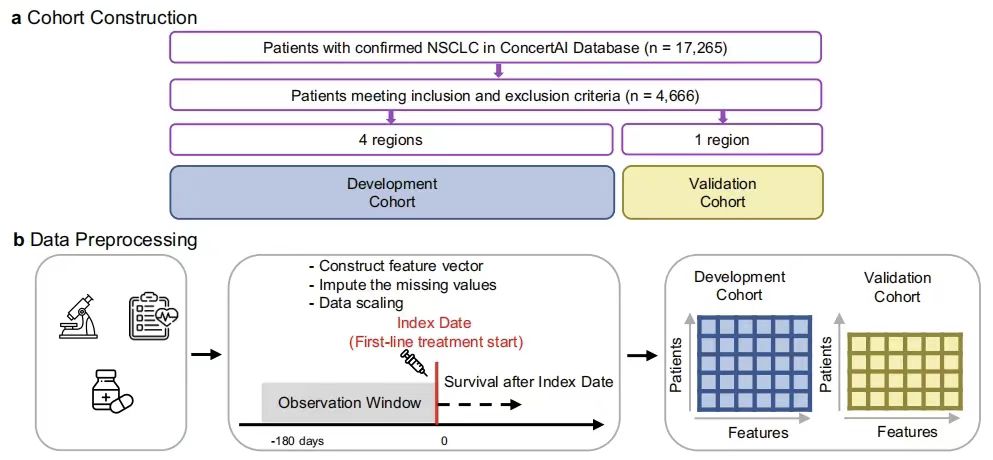
Constructing a cohort of patients with advanced non-small cell lung cancer based on ConcertAI's large-scale real-world dataset
The study used the ConcertAI Patient360™ NSCLC dataset from the US Oncology Electronic Health Record (EHR) database to construct a cohort of patients with advanced non-small cell lung cancer (aNSCLC) receiving first-line (1 L) immune checkpoint inhibitor (ICI) treatment.This dataset is a de-identified, patient-level dataset based in the United States, extracted from the ConcertAI network, covering more than 8 million unique patients.Data from more than 900 oncology and hematology cancer clinics, representing patients treated in community and academic practices across all 50 states, were extracted, including data on disease recurrence date and type, histology, PD-L1 testing information, tumor response, ECOG-PS, and comorbidities.
As shown in the figure below, this study selected patients with histologically confirmed non-small cell lung cancer (NSCLC) from January 2015 to January 2023 (n=17,265) to construct a retrospective, observational cohort. After the inclusion/exclusion criteria and the exclusion of patients without valid overall survival (OS) records,4,666 patients were included in the study, and the patients were represented by a 104-dimensional vector, with dimensions including demographic information, laboratory tests and other variables.
Based on the geographic regions of clinical institutions defined by the U.S. Census Bureau, the researchers divided the cohort into model development (Northeast, South, and West regions, n=3,225) and validation subcohorts (Midwest region, n=1,441). The two have similar demographics, and the validation subcohort has a higher proportion of white patients and patients in community medical institutions. The observation period of the study was 180 days before the index date, and overall survival (OS) was defined as the time from the index date to death due to any cause, and progression-free survival (PFS) was defined as the time from the index date to the first real-world progression event or death due to any cause. The purpose is to solve problems such as the prediction of survival in patients with advanced non-small cell lung cancer through relevant analysis of this data set.
GEMS model construction: GNN-based identification of survival subphenotypes and prediction performance validation for advanced non-small cell lung cancer
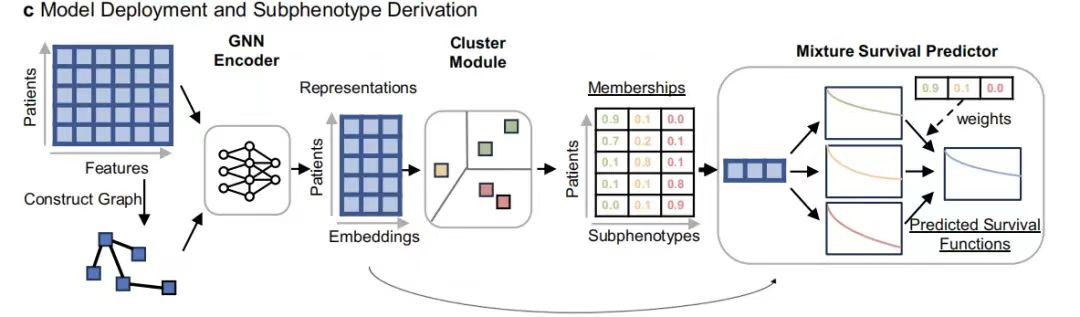
In this study, the GEMS model was designed to identify predictive subphenotypes associated with real-world overall survival (OS) characteristics in patients with advanced non-small cell lung cancer (aNSCLC).Its core architecture includes GNN Encoder, Cluster Module and Mixture Survival Predictor.
Among them, the GNN encoder effectively extracts high-order patient representations by capturing the graph structure relationship of the patient's 104-dimensional feature vector (covering variables such as demographics, laboratory tests, and metastasis status); the encoded representations are input into the clustering module to generate sub-phenotypes with survival prediction value as the basic components of the hybrid model.
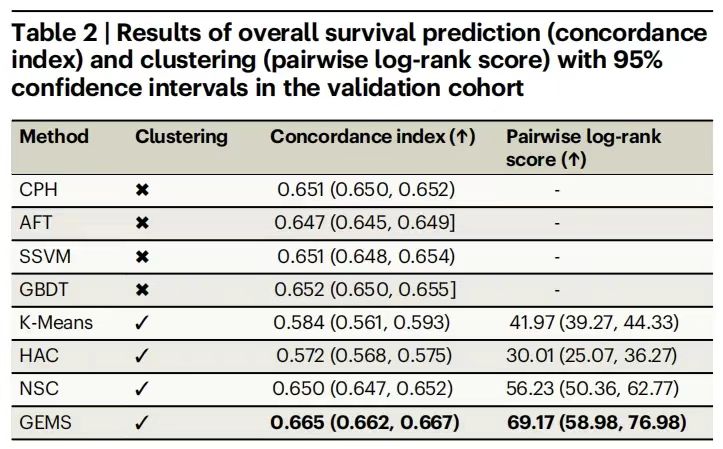
The model training first used the development cohort (n=3,225) as data support, and used the consistency index (c-index) and pairwise log-rank score as evaluation indicators, and compared them with traditional baseline models such as Cox proportional hazard regression (CPH), gradient boosted decision tree (GBDT), neural survival clustering (NSC) and unsupervised methods such as K-means and hierarchical clustering.
The experimental results are shown in the following table.GEMS performed well in predicting overall survival.The average c-index reached 0.665 (95% CI: 0.662-0.667), significantly higher than the best baseline model GBDT's 0.652; the log-rank score was 69.17 (95% CI: 58.98-76.98), far exceeding NSC's 56.23, verifying the effective use of data features by the supervised learning framework.
Then,This study further characterized the impact of the GNN encoder on GEMS by visualizing the representations derived from patients and their GNN encoders.The Uniform Manifold Approximation and Projection (UMAP) is used. As shown in the figure below, through the visualization of the Uniform Manifold Approximation Projection (UMAP), it is found that in the patient representation space output by the GNN encoder, the patient groups with different total survival times are clearly separated, while the various types of patients in the original feature space are mixedly distributed, which intuitively reflects the modeling ability of the graph neural network for complex feature relationships.
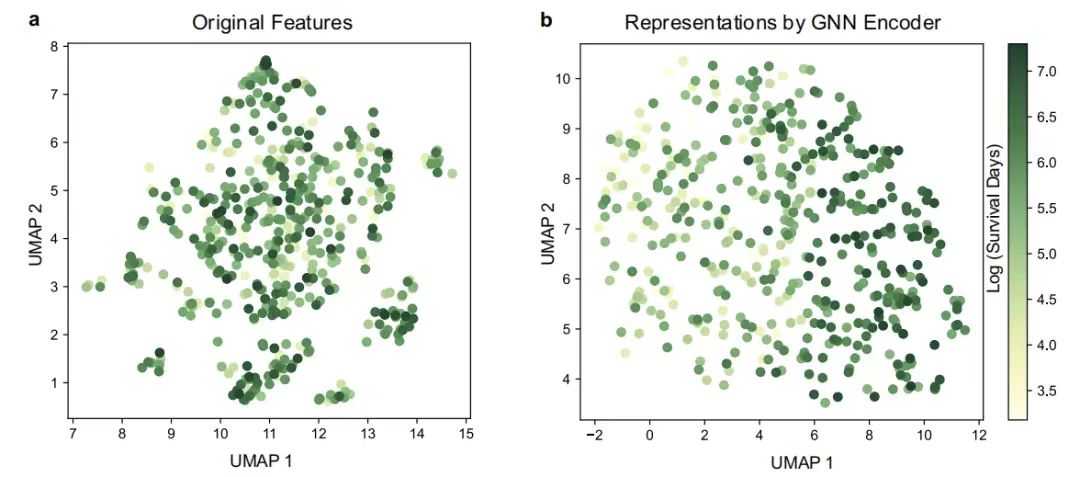
UMAP visualization of patients
Figure a: UMAP visualization of original features; Figure b: UMAP visualization of features obtained by GNN encoder
As shown in the figure below,The researchers further used the model to identify three predictive subphenotypes with significant survival differences:Subphenotype 1 (n=1335) was characterized by a high proportion of females (55.50%), mild comorbidities, and low metastatic burden, with an average overall survival of 688 days, and the lowest use rates of cough suppressants, β-blockers, and the incidence of bone/brain/adrenal metastases. The survival curve of subphenotype 2 (n=420) showed a mid-term risk increase, with intermediate comorbidities and metastatic burden. Subphenotype 3 (n=1420) had a female proportion of 35.21% and an average overall survival of only 321 days, characterized by multiple medications, a high metastatic rate (liver metastasis 31.20%, bone metastasis 51.48%) and severe comorbidities (water and electrolyte disorders 8.31%, kidney abnormalities 21.43%), and the most complex co-occurrence pattern of metastasis-comorbidities-laboratory abnormalities.
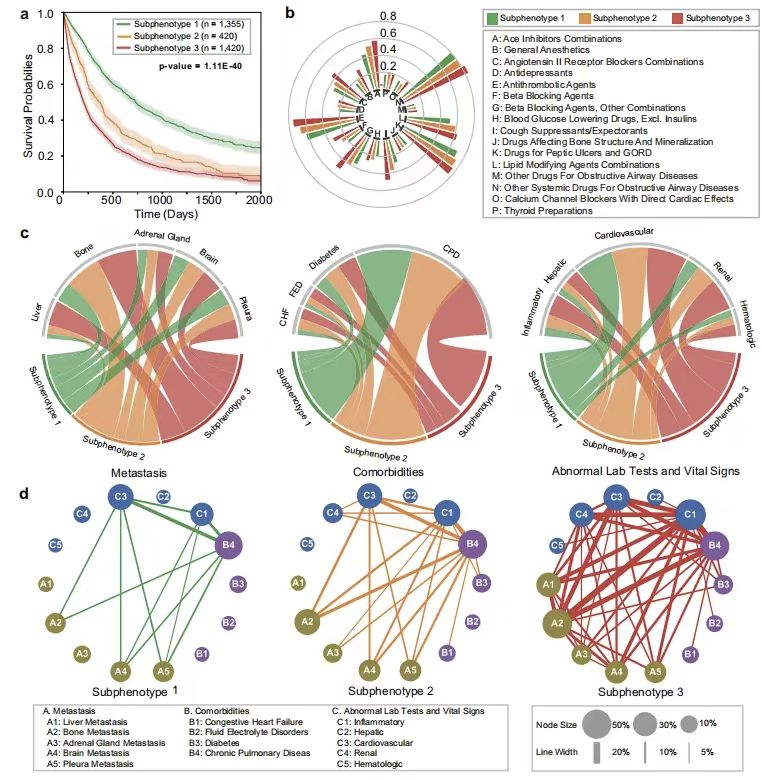
* Figure a: Kaplan-Meier curves of overall survival for each subphenotype
* Figure b: Sunburst diagram of drug administration rate of each subtype
* Figure c: Chord diagram of differences in classification of metastasis (left), comorbidities (middle), and abnormal clinical features
* Figure d: The incidence of different sub-phenotypes
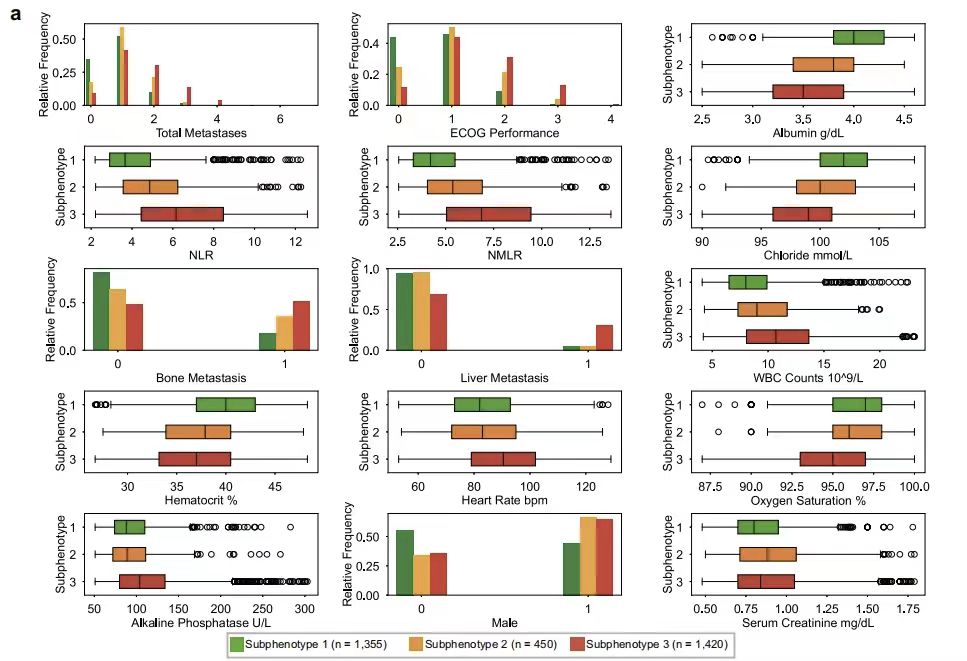
In order to further understand the different characteristics between different subphenotypes, the study tested the differences in each variable between each subphenotype. As shown in the figure below, the key predictor analysis showed that the Eastern Cooperative Oncology Group performance status (ECOG Performance) and the total number of metastatic sites (Total Metastases) are the core indicators for distinguishing subphenotypes. In terms of laboratory indicators, the neutrophil-to-lymphocyte ratio (NLR) and the neutrophil-to-monocyte-to-lymphocyte ratio (NMLR) are characteristic parameters of subphenotype 2, while subphenotype 1 is associated with normal albumin levels (WBC Counts) and high hematocrit (Hematocrit), and subphenotype 3 is closely associated with increased heart rate (Heart Rate bpm), decreased oxygen saturation (Oxygen Saturation), and increased alkaline phosphatase (Alkaline Phosphatase).
The above results show thatThe GEMS model not only achieves accurate stratification of the survival prognosis of aNSCLC patients,Furthermore, through the analysis of sub-phenotype characteristics, it provides a clinical decision-making basis based on real-world data for the formulation of individualized treatment strategies.
The global revolution in precision diagnosis and treatment of lung cancer: How do AI and multi-omics technologies change the survival landscape?
In the field of lung cancer diagnosis and treatment, a revolution driven by artificial intelligence (AI) and precision medicine is reshaping clinical practice. A research team at the University of Toronto in Canada has developed an AI-assisted blood test technology that analyzes EGFR mutations in circulating tumor DNA.Combining machine learning with clinical data effectively improves the recognition rate of people who benefit from targeted treatment.It enables patients carrying EGFR sensitive mutations to accurately receive EGFR tyrosine kinase inhibitor (TKI) treatment, significantly prolonging the median progression-free survival.
Paper link:https://pubmed.ncbi.nlm.nih.gov/35624472/
The "evA.I. system" of University College London uses 27-dimensional clinical data.Accurately predict immune checkpoint inhibitor (ICI) responses and help identify drug-resistant populations.Thereby improving the effectiveness of immunotherapy and prolonging the median overall survival.
Paper link:https://pmc.ncbi.nlm.nih.gov/articles/PMC10957591/
In China, innovative achievements from universities and enterprises continue to emerge in the research of precision diagnosis and treatment of advanced non-small cell lung cancer.,Professor Zhang Peng's team from Tongji University and the Chinese Academy of Sciences team completed the first international small cell lung cancer protein genomics map study,By integrating the multidimensional omics data of 112 samples, we found that high expression of HMGB3 protein was associated with poor prognosis, and established an immunotherapy benefit prediction model based on ZFHX3 mutation status, opening up a new path for precision treatment guided by molecular typing.
Paper link:https://doi.org/10.1016/j.cell.2023.12.004
Tsinghua University Shenzhen International Graduate School and Shenzhen People's Hospital jointly developed the "AI + Intelligent Pathology" system.After deep learning more than 3,000 difficult cases, it can accurately identify the histological types of poorly differentiated lung cancer with an accuracy rate of 97%.Shorten the decision-making cycle of targeted treatment. The AI prediction model based on blood glycoprotein markers by the team can warn of lung cancer risk 3 years in advance, with a clinically verified accuracy rate exceeding 92%, providing a non-invasive solution for ultra-early screening.
Paper link:https://www.nature.com/articles/s41598-025-98731-4
Reference articles:
1.https://mp.weixin.qq.com/s/LBcVbQUpTYRnKZ5I1KY_VA