Command Palette
Search for a command to run...
Published in Nature, Russian Research Team Uses Machine Learning to Search Trillions of Mass Spectrometry Data and Discover Unknown Chemical Reactions

Mass spectrometry (MS) is one of the core technologies of modern chemical research. By measuring the mass-to-charge ratio (m/z) of molecular ions, mass spectrometry can provide key information about the molecular formula, structure and even reaction mechanism of a compound. The emergence of High-Resolution Mass Spectrometry (HRMS) has increased the accuracy of analysis to the part per million (ppm) level, becoming the "gold standard" in organic synthesis, metal catalysis, drug development and other fields. However, with the improvement of the degree of instrument automation, the amount of mass spectrometry data generated by laboratories every day has exceeded the terabyte (TB) level, resulting in TBs of information piling up on computers. But currently,Experimental and MS data rely heavily on manual analysis, and human factors can affect the interpretation coverage of data analysis.This severely limits the experiment.
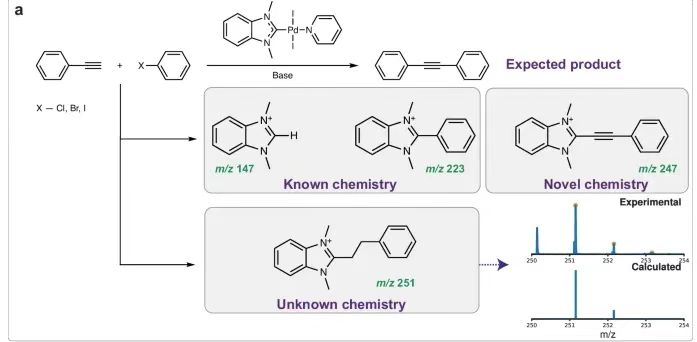
To address this challenge, researchers from the Russian Academy of Sciences and other institutions introduced an innovative machine learning (ML) driven search engine MEDUSA Search.Ion isotope distribution can be searched in multi-component high-resolution mass spectrum databases up to TB level.This method uses an isotope distribution-centric search algorithm and is enhanced by two collaborative machine learning models to assist in the discovery of unknown chemical reactions. This method can rigorously screen existing data to provide effective support for chemical hypotheses while reducing additional experiments. In addition, as an extension of the baseline method, the model can automatically generate reaction hypotheses and reveal new chemical transformations. Among them,The heterocycle-vinyl coupling process in the Mizoroki-Heck reaction stood out in the experiments, highlighting the engine’s ability to resolve complex chemical phenomena.
The related research, titled "Discovering organic reactions with a machine-learning-powered deciphering of tera-scale mass spectrometry data", has been published in Nature Communications.
Research highlights
* Mining unknown reactions: Instead of relying on new experiments, use existing data to mine unknown chemical reactions, reducing experimental costs and resource consumption.
* Efficient search algorithm: A unique isotope distribution search algorithm combined with a machine learning model can accurately search for ions in large-scale mass spectrometry data and reduce misjudgment.
* Expand chemical cognition: Discover new reaction pathways and products, such as the heterocyclic-vinyl coupling process in the Mizoroki-Heck reaction, and deepen the understanding of chemical reactions.
Paper address:
The open source project "awesome-ai4s" brings together more than 100 AI4S paper interpretations and provides massive data sets and tools:
https://github.com/hyperai/awesome-ai4s
Dataset: Over 20,000 mass spectrometry images, confirming the presence of reactive ions
Since most mass spectrometry signals lack professional analysis, the laboratory has accumulated and stored a huge amount of data over the past few years. The data used in this experiment all come from this data. These mass spectrometry data cover many chemical transformation studies.The total data volume exceeds 8 TB, including more than 20,000 mass spectra.Multi-component high-resolution mass spectrometric data at different resolutions are stored, enabling confirmation of the presence of target ions in a wide range of applications.
MEDUSA Search During the reaction discovery process, the generated ion formulas are searched against the entire terascale HRMS database to find new reaction pathways and products, and the data is visualized.
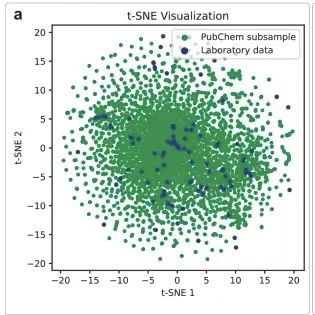
The dataset was visualized using the t-SNE dimensionality reduction technique. To demonstrate the high diversity of the archived dataset, the researchers created two t-SNE plots.The molecules collected were randomly sampled from the PubChem database and mass spectrometry registered compounds.The compounds registered in the analytical mass spectra cover the chemical space very well. Each point represents a spectrum, similar mass spectra are close to each other on the graph, and different workers recorded different spectra for comparison.The compounds in the mass spectra are widely distributed in the chemical space, and the mass spectra recorded by different researchers vary greatly.As shown below.
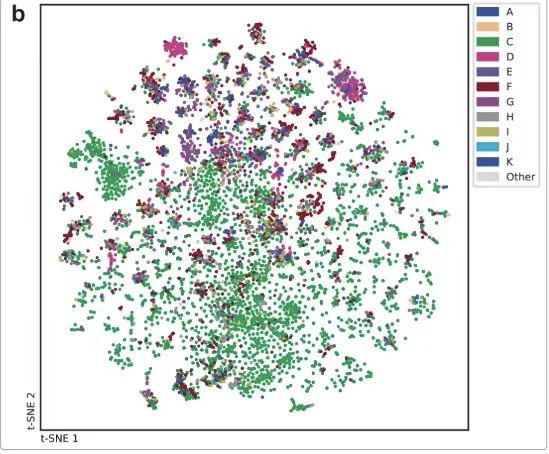
The diverse data generated by the study has been stored on Figshare.This contains a 9 GB mass spectrometry ZIP archive,All discovered products mentioned are included, and additional reaction mass spectrometry data are included to test the search engine functionality. Some data that did not yield results cannot be shared publicly due to confidentiality or intellectual property reasons.
* figshare is an online data repository based on cloud computing technology, where researchers can save and share their research results, including data, datasets, images, videos, posters and codes.
HRMS High Resolution Mass Spectrometry Dataset:
Model architecture: Discovering unknown chemical reactions based on isotope distribution searches
MEDUSA Search is a machine learning-based mass spectrometry data analysis engine that can be used to discover unknown chemical reactions from massive mass spectrometry data.
Specifically, the search process developed in MEDUSA Search consists of 5 steps.
first,MEDUSA Search takes as input the molecular formula and charge of the ion being searched for.These molecular formulas or charges can be derived from the reaction system using hypothesis generation methods or can be defined manually (as shown in Figure A below). The search engine then searches all spectral files that contain the two most abundant isotopologue peaks of the input ion, as shown in Figure B below. The isotopologue peaks are represented by their mass-to-charge ratio, m/z. These spectral files are called candidates, and the researchers also perform a cosine distance threshold calculation for the spectral files, as shown in Figure C1 below. Next, an algorithm is executed on all candidate mass spectra that searches for isotopic distributions according to the input formula within a single spectrum, as shown in Figure C2 below.
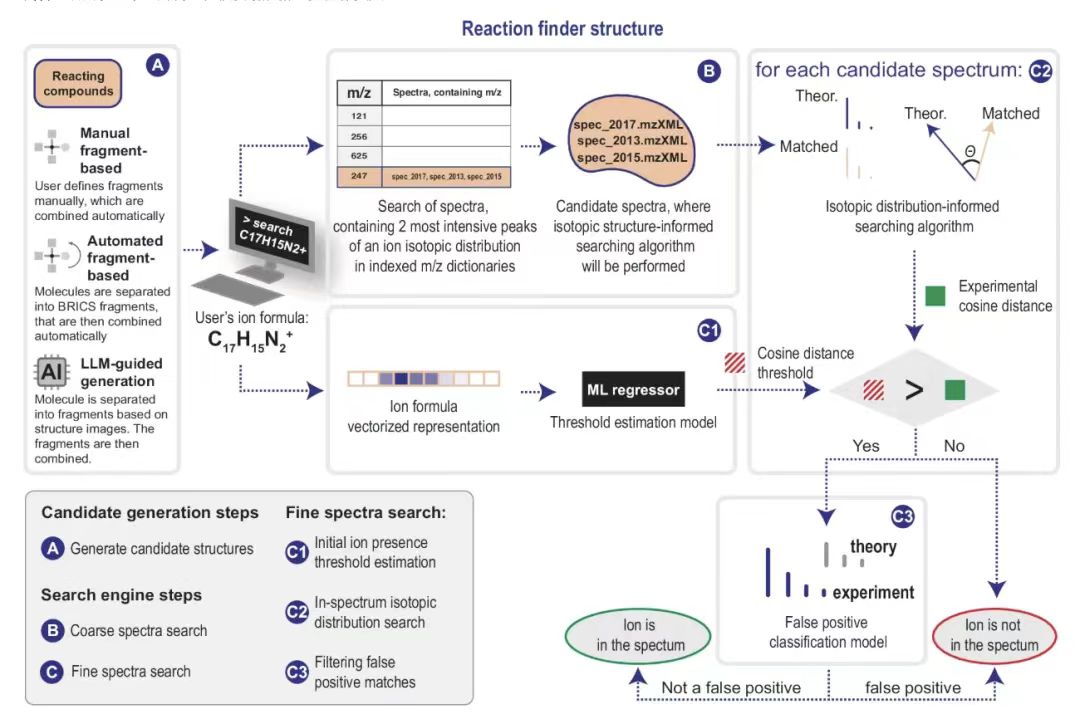
Before searching, researchers shouldGenerate a list of hypothetical reaction pathways based on prior knowledge of the reaction system(Figure A). This system is designed around the recombination of breakable bonds and the corresponding fragments. Information about the chemical formula and charge is input, and the theoretical "isotope pattern" of the ion can be calculated. The two most abundant isotopologue peaks are searched in the inverted index (Figure B). Mass spectra containing these peaks are called candidates. After the coarse spectrum search, each candidate spectrum is searched for the isotope distribution of the query ion.There are 3 steps involved:
* Initial ion presence threshold estimate:The cosine distance returned by the isotope distribution search algorithm within the spectrum is used as a measure of the similarity between the theoretical and matched isotope distributions. The automatic determination of the presence of an ion in the spectrum depends on the estimated maximum cosine distance (i.e., the ion presence threshold). Based on a machine learning regression model (Figure C1), the ion presence threshold is determined using the input ion formula.
* Search for isotope distribution within a spectrum:The intraspectral isotope distribution search algorithm (Figure C2) matches the peaks in the experimental candidate mass spectrum with the peaks in the theoretical isotope distribution; the cosine distance is calculated at each step to select the most similar peak. If no peak is found, it is replaced with a peak with an intensity equal to the median noise. If the final cosine distance is less than the ion presence threshold estimated in step (Figure C1), the ion is considered to have been found.
* Filter false positive matches:Additional machine learning classifiers (Figure C3) use information about neighboring peaks to detect false positive ions. This problem often manifests itself as searching for a distribution that is part of another distribution. One of the most prominent examples starts with M+1, where M also exists.
Experimental conclusion: Heterocyclic-vinyl coupling experiments highlight the model detection capabilities
The 520 generated ions were searched through the entire Terascale HRMS database with a total computational time of 3–4 days (8–11 min per ion). Experimental results show that MEDUSA Search detects multiple isotope distribution patterns.
The formation of catalytic conversion products is closely related to the corresponding reaction mechanism.Previous researchers have conducted several Mizoroki-Heck and cross-coupling reactions (such as Sonogashira, Suzuki, Buchwald-Hartwig, etc.), in which the catalytic components are Pd/NHC complexes with different NHC ligands and halogen substituents. In the process of studying the reaction mechanism through ESI-MS spectra of the reaction mixture, the coupling products [NHC-H]⁺, [NHC-Ph]⁺, [NHC-O]⁺ and [NHC-N]⁺ were found. Based on these observations,The key roles of R-NHC coupling and M-NHC bond cleavage in the evolution of M/NHC complexes under catalytic reaction conditions were revealed.The formation of catalytically active molecular M/NHC catalysts and “NHC-free” cocktail-type catalysts is described from the perspective of the number of CC coupling reactions, including H-NHC salt and O-NHC coupling formation.
In the Sonogashira reaction, a previously unknown ethynyl-NHC coupling product was isolated and a possible reaction pathway was described. The ethynyl-NHC coupling product is very reactive and can undergo various transformations. The hydrogenated derivative of the product was analyzed using the described method.The ESI-MS spectrum of the Sonogashira reaction mixture showed the presence of [NHC-(CH₂)₂-Ph]⁺ product.As shown below. It is speculated that this process occurs through a transfer hydrogenation reaction.
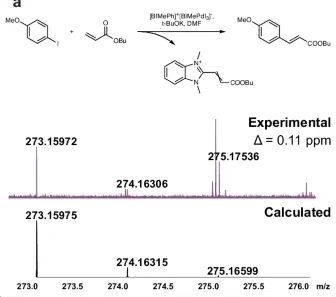
Under the catalysis of Pd/NHC complex [BIMePh]⁺ [BIMePdI₃]⁻,Mass spectrometry analysis of the Mizoroki–Heck reaction mixture between p-methoxyiodobenzene and butyl acrylate revealed the formation of [BIMe (CH)₂COOBu]⁺.The molecular formula was confirmed by ultra-high resolution mass spectrometry. Experiments involving the formation of [IPrCHC(Ph)COOBu]⁺ were used to distinguish between homogeneous and heterogeneous catalysis by mercury. The interference of mercury on the reaction species was eliminated and other conditions were kept the same as the original experiments. The molecular formula was also confirmed by ultra-high resolution mass spectrometry and the chemical structure was verified by MS/MS experiments.
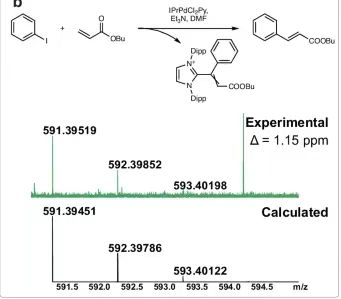
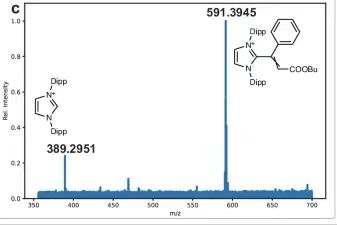
Experiments were performed with five different NHC ligands. The possibility of vinyl-NHC coupling during Pd/NHC transformations in the Mizoroki–Heck reaction was tested. Vinyl-NHC products were found in all cases studied, independent of the ligand in the complex, and all products were defined with very small errors. For the reaction mixtures studied, such as (BIMe)PdI₂Py, (SIMes)PdCl(allyl), and (PIPr)PdCl(allyl),In addition to vinyl-NHC, ethyl-NHC was also detected.The m/z errors for the (IMes)PdCl(allyl) and (SIPr)PdCl(allyl) complexes were very low, less than 0.3 ppm, while the errors were below 1 ppm. In all MS experiments, the configuration was set to prevent transitions from occurring during the recording of the mass spectra. Pressure sample infusion ESI-MS reaction monitoring was also performed for the vinyl-NHC coupling process in question to confirm that ions could be observed in multiple modes of reaction data collection.
This robust machine learning-based reaction discovery computational engine has been demonstrated to be able to use ions of various compositions.Ion searches can be performed on all MS instruments at resolutions sufficient to observe isotopic distributions.Combining the developed system with other computational techniques (e.g., algorithms for predicting ion fragments by structural formula or peptide sequence, different adduct calculators) could become a powerful analytical tool for comprehensive screening, which is essential to accelerate discoveries in various scientific fields.
also,This method also realizes the research concept of "Experimentation in the Past".It fully taps into the value of existing data, discovers new reaction pathways and products, saves research resources, provides new ideas and methods for chemical research, and promotes the development of the field of organic chemistry. In terms of practical applications, it can help pharmaceutical companies, material research and development companies, etc. find new reaction pathways and products faster, reduce research and development costs, improve research and development efficiency, and provide a powerful analytical tool for chemical research.
Automated analysis of mass spectrometry data enters clinical application
With the continuous deepening of mass spectrometry technology in scientific research and industrial production, automation technology has begun to move towards clinical application. As an important component of precision diagnostic technology, clinical mass spectrometry can achieve full automation from sample collection, processing, separation to analysis. According to the 17th edition of the Global IVD Industry Report newly released by the United States, the market size of the global clinical mass spectrometry industry will be US$930 million in 2024.It is expected to reach US$1.435 billion in 2029. From 2024 to 2029, the clinical mass spectrometry market is expected to grow at an average annual rate of 9%, becoming the fastest growing market segment in the IVD field after nucleic acid testing.
* IVD (in vitro diagnostic products) refers to medical devices, in vitro diagnostic reagents and drugs.
Looking at the Chinese market,The clinical mass spectrometry industry has long entered the fast lane of development, with significant progress in mass spectrometry multi-omics, domestic mass spectrometers and automated mass spectrometry.According to the "2024 Clinical Mass Spectrometry Industry Research Report", as of July 31, 2024, excluding quality control products and calibration products, a total of 228 domestic clinical mass spectrometry products have been approved by NMPA.
In terms of the types of approved reagents, the number of domestically produced clinical mass spectrometers approved in China has been growing in the past five years, and there has been no sign of slowing down. As of July 31, 2024, 51 reagents have been approved for vitamin testing, 46 reagents have been approved for drug concentration monitoring, and 45 reagents have been approved for chronic diseases and hormones. From 2020 to 2023, there will be 10, 12, 13, and 16 reagents respectively.
Among the approved instruments, liquid chromatography-mass spectrometry (LC-MS) instruments are the main ones, with a total of 33 Chinese-made LC-MS devices approved. The second is domestic matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF MS) devices, with a total of 25 approved, which are approved for microbial detection, nucleic acid detection and peptide detection.
* Liquid chromatography-mass spectrometry is an analytical chemistry technique that combines the physical separation capabilities of liquid chromatography (LC) with the mass analysis capabilities of mass spectrometry (MS).
* Matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF MS) is a new type of soft ionization biomass spectrometry developed in recent years and is widely used to identify a large number of bacteria and fungi.
At present, the clinical application of LC-MS in China has been relatively new and is still in its infancy. There are still many deficiencies. Many factors, such as IVD manufacturers, medical testing laboratories, professional and technical personnel, management departments and policies, may affect the application of clinical mass spectrometry detection technology. However, looking forward to the future, the combination of automation and intelligence is bound to be an important development direction. The clinical application of LC-MS/MS will continue to develop, and while improving the detection efficiency and accuracy, it will further help doctors interpret the results and assist clinical decision-making.
References:
1.https://mp.weixin.qq.com/s/27drrM5lwawHRgRMWvHZRQ
2.https://mp.weixin.qq.com/s/pkd2I573on08syPkqdStOQ