Command Palette
Search for a command to run...
Selected for CVPR 2025, the Harbin Institute of Technology Team Proposed a Hierarchical Distillation multi-instance Learning Framework HDML to Quickly Process Gigapixel Pathology full-slice Images

Pathological images contain rich phenotypic information, and pathological diagnosis based on pathological images is widely regarded as the "gold standard" for cancer diagnosis. Among them, the whole slide image (WSI) is a high-resolution digital pathological image that uses full-slide digital scanning technology to convert pathological tissue slices into digital images up to 1 billion pixels. It has the characteristics of high resolution, panoramic display, and large data volume. It is the mainstream method of current medical diagnosis and medical research.
Multi-Instance Learning (MIL) is one of the main methods for analyzing WSI, and has achieved good performance in tasks such as tumor detection, tissue microenvironment quantification, and survival prediction. However, since WSI contains a huge amount of information, using MIL for reasoning faces the challenge of high costs. The first is the data preprocessing problem. The WSI cropping and feature extraction process is very time-consuming; the second is the redundant patch problem. WSI usually contains redundant patches, which contribute the least to bag-level classification. The simplest way to solve the above problems is to eliminate irrelevant examples through attention scores. However, the existing MIL algorithm needs to extract the features of all cropped blocks before calculating the attention score, which undoubtedly creates a "chicken and egg" problem.
Based on the above analysis, Professor Jiang Junjun, Associate Professor Jiang Kui from Harbin Institute of Technology, and Professor Zhang Yongbing from Harbin Institute of Technology (Shenzhen) in China innovatively demonstrated a solution to reduce inference time. The team proposed a hierarchical distillation multi-instance learning framework (HDMIL), which aims to quickly identify irrelevant patches, thereby achieving fast and accurate classification. From the experimental results, compared with previous advanced methods, HDMIL reduced the inference time on 3 public datasets by 28.6%.
The related results were published under the title "Fast and Accurate Gigapixel Pathological Image Classification with Hierarchical Distillation Multi-Instance Learning" and were selected for CVPR 2025.
Research highlights:
* The proposed method speeds up the reasoning process while also improving classification performance, achieving a balance between speed and performance that traditional methods cannot achieve, and provides inspiration for future research on multi-instance classification
*This method demonstrated for the first time the Kolmogorov-Arnold classifier based on Chebyshev polynomials and applied it to digital pathology, significantly improving the classification performance
* The proposed method has been verified by a large number of experiments and has achieved reliable and effective verification results on 3 public datasets

Paper address:
https://arxiv.org/abs/2502.21130
Dataset: Three major public data sets verify effectiveness
To ensure the effectiveness of the experiment, the researchers evaluated the effectiveness of the proposed method on three public datasets:
* Camelyon16 dataset is used for breast cancer lymph node metastasis detection, where the ratio of training set and validation set is divided according to the official training set 9:1, and the official test set is used for testing across all folds
*The TCGA-NSCLC dataset was used for lung cancer classification. The dataset was divided into training set, validation set, and test set in a ratio of 8:1:1.
*Using the TCGA-BRCA dataset for breast cancer subtype classification, the ratio of training set, validation set and test set is also 8:1:1
It is worth noting that all WSIs were preprocessed using the tools developed by CLAM, and the experiments followed 10-fold Monte Carlo cross-validation.
Model architecture: Two-stage architecture involves training and reasoning, and innovatively introduces the Kolmogorov-Arnold classifier
The HDMIL framework proposed by the institute involves two parts: training and reasoning. In this framework, there are two key components: the dynamic multi-instance network (DMIN), which is designed to classify high-resolution WSI and identify instances that are not related to bag-level classification; the lightweight instance prescreening network (LIPN), a network specially tailored for low-resolution WSI.
Before training, the researchers first preprocessed the input data following the standard procedure for pathological WSI. The dataset consists of S WSI pyramids with slide labels, each Xᵢ containing a pair of high-resolution (20x) and low-resolution (1.25x) WSIs, denoted as Xᵢ,ₕᵣ and Xᵢ,ₗᵣ, respectively.
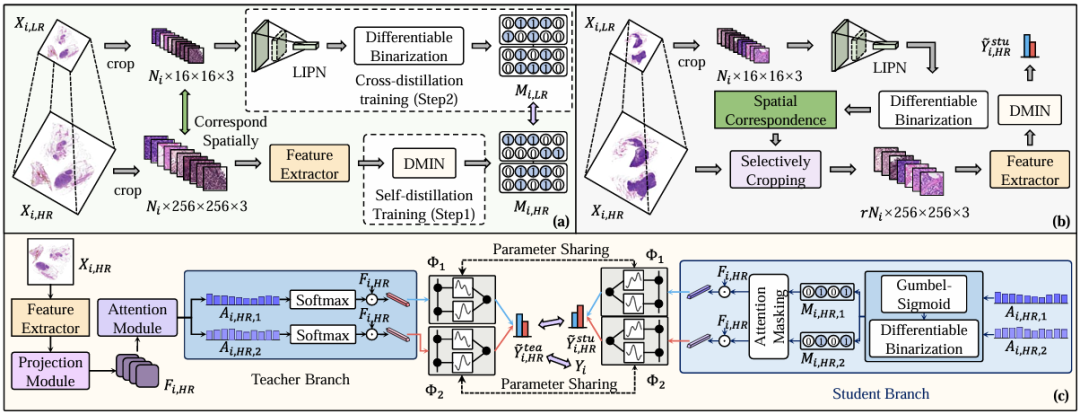
Specifically, Figure a shows the training phase, as shown in the figure below. The researchers first used a self-distillation training strategy to train DMIN using high-resolution WSI (Xᵢ, ₕᵣ) to enable it to perform bag-level classification and indicate irrelevant areas. Although DMIN successfully identified irrelevant areas in WSI, it did not improve the inference speed. This is because DIMN needs to use the features of all patches generated by the feature extractor to determine which instances should be eliminated, and patch-wise feature extraction is actually the key to breaking the bottleneck of WSI inference speed.
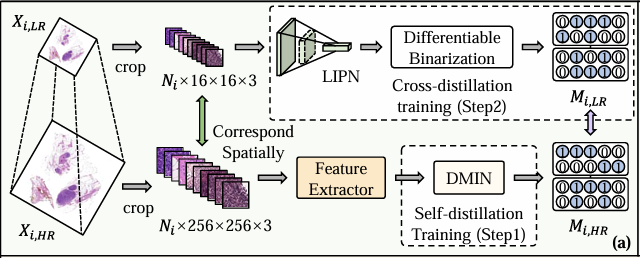
Therefore, the researchers then freeze the DMIN and use the generated mask to extract the LIPN. As mentioned above, LIPN is a lightweight instance pre-screening network tailored for low-resolution WSI, which is trained by cross-distillation using low-resolution WSI (Xᵢ,ₗᵣ) and can quickly identify irrelevant regions in low-resolution WSI, thereby indirectly indicating irrelevant patches in high-resolution WSI.
In terms of specific implementation, the researchers used the widely used ResNet-50 as a feature extractor, which is a model pre-trained on ImageNet, and used a lightweight variant of MobileNetV4 for the pre-screening network LIPN. Through the above steps, the researchers achieved the discrimination of the binary importance (important or not) of each region at a very low computational cost.
Figure c shows the self-distillation training of DMIN on high-resolution WSI (Xᵢ, ₕᵣ), as shown in the figure below. It can be seen that DMIN consists of 5 modules, including the Projection Module, the Attention Module, the Teacher Branch, the Student Branch, and the CKA classifiers.
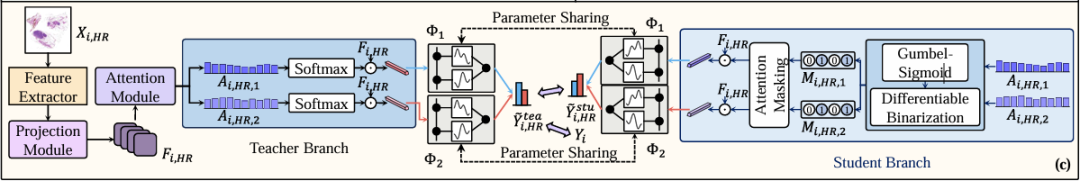
Specifically, all patches extracted from the high-resolution WSI (Xᵢ,ₕᵣ) are first input into the pre-trained feature extractor to generate a set of example-level features Iᵢ,ₕᵣ, which are then input into the projection module for dimensionality reduction to obtain a new feature set Fᵢ,ₕᵣ, which are then input into the attention module to calculate the unnormalized attention score.
In the teacher branch, the reduced Fᵢ,ₕᵣ is linearly weighted using the attention matrix of each category to produce a bag-level representation for the final classification. Only a subset of examples with larger attention scores are used in the student branch to calculate the bag-level representation, and the researchers also impose constraints to ensure that its bag-level representation is as consistent as possible with the representation obtained in the teacher branch using all instances. Through this method, the attention module is made to pay more attention to instances that are more important for bag-level classification and filter out irrelevant instances. At the same time, the optimization process also uses the Gumbel trick to selectively use instances with higher attention scores for end-to-end training to avoid the occurrence of non-differentiable problems.
Finally, in order to enhance the ability of the MIL classifier, the researchers proposed using the Kolmogorov-Arnold network to learn nonlinear activation functions instead of using fixed activation functions in the classifier. And by designing a hybrid loss function, the researchers facilitated the achievement of three training goals of DMIN. The first is that the teacher branch can correctly classify Xᵢ,ₕᵣ, the second is that the use of some instances in the student branch can be consistent with the classification results of using all instances in the teacher branch, and the third is that the proportion of selected instances should be controllable.
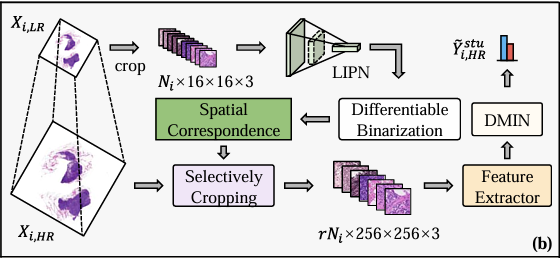
Figure b shows the inference phase, as shown in the figure below. The specific process can be divided into three steps: the first step is to crop all patches in the low-resolution WSI (Xᵢ,ₗᵣ), with a total of Nᵢ; the second step is to input these patches into LIPN to identify classification-related areas and generate Mᵢ,ₗᵣ; the third step is to selectively crop the corresponding patches in Xᵢ,ₕᵣ based on Mᵢ,ₗᵣ, and then input the remaining patches into the feature extractor and DMIN, and finally calculate through the cross-category student branches to generate the final classification results.
Research results: "Simplified" HDMIL still outperforms existing advanced methods
Based on the three datasets of Camelyon16, TCGA-NSCLC, and TCGA-BRCA, the researchers compared the classification performance of HDMIL with 11 MIL methods, including Max-pooling, Mean-Pooling, ABMIL, CLAMSB, CLAMMB, DSMIL, TransMIL, DTFDAFS, DTFDMAS, S4MIL, and MambaMIL.
It is worth mentioning that the researchers tested different configurations of HDMIL, namely HDMIL† and HDMIL. The former means that only DMIN is used for reasoning without pre-screening instances through LIPN. The specific results are shown in the figure below:
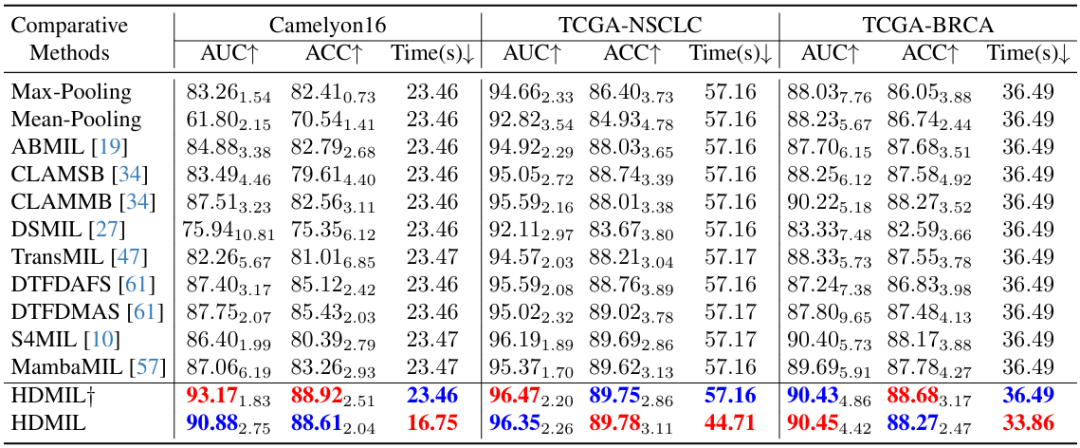
It can be seen that both HDMIL† and HDMIL have consistently better test results on the three datasets than existing methods. For example, on the Camelyon16 dataset, HDMIL achieved an AUC of 90.88% and an accuracy of 88.61%, which are 3.13% and 3.18% higher than the previous best method, respectively.
At the same time, when the dataset is large enough, HDML improves the speed without affecting the classification performance. For example, on TCGA-NSCLC and TCGA-BRCA, both contain about 1,000 WSIs, but the difference in test performance between HDML† and HDML is not large, proving that HDML has achieved an excellent balance between inference speed and classification performance.
In addition, HDMIL† is on par with other existing methods in terms of processing time, while HDMIL is significantly superior to all methods. This is because HDMIL†, like other methods, needs to process the same number of high-resolution patches. HDMIL reduces the time spent on this data processing through LIPN, thus significantly reducing the time spent on inference compared to other methods on the three datasets, achieving speed increases of 28.6%, 21.8%, and 7.2% respectively.
In order to analyze the impact of each component, the researchers conducted an ablation experiment to further illustrate the impact of each module in HDMIN on the classification results, as shown in the figure below. The results show that the proposed CKA classifier replaces the traditional linear layer-based classifier and incorporates self-distillation into DMIN training, which significantly improves the classification performance.
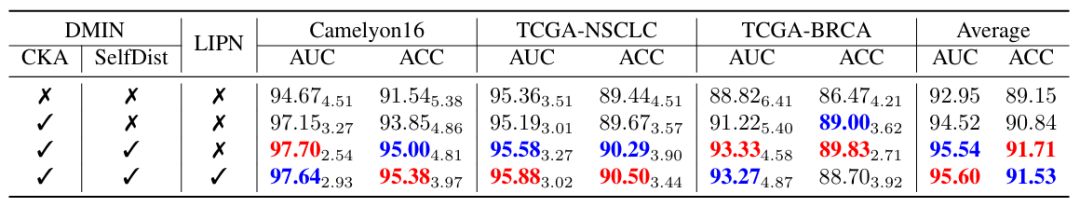
In general, the proposal of HDMIL is undoubtedly a new idea and attempt. The feasibility of its idea has been proved through a large number of experiments. It provides a new method for analyzing pathological images, especially WSI, using the MIL method, and accelerates the vigorous development of digital pathology.
Digital pathology thrives with AI
In recent years, the booming development of digital pathology has led to a new round of medical and biological progress, especially in the fight against cancer, one of the biggest enemies of mankind. It is worth mentioning that the proposal of HDMIL is not the first attempt of the Harbin Institute of Technology team in this field.
Last year, CVPR 2024 included a study titled "Virtual Immunohistochemistry Staining for Histological Images Assisted by Weakly-supervised Learning". The article mentioned a weakly supervised learning method called confusion-GAN for virtual immunohistochemistry (IHC) staining, which can convert H&E images into IHC images, solving the cumbersome and expensive cost of traditional methods in IHC staining.

Paper address: https://openaccess.thecvf.com/content/CVPR2024/papers/Li_Virtual_Immunohistochemistry_Staining_for_Histological_Images_Assisted_by_Weakly-supervised_Learning_CVPR_2024_paper.pdf
In addition to the same authors as the above-mentioned research, this article was also co-authored by Professor Jiang Junjun and Professor Zhang Yongbing, which further confirms Harbin Institute of Technology's deep cultivation and accumulation in this field.
Of course, as the two corresponding authors of the article, Professor Jiang Junjun and Professor Zhang Yongbing are also worth highlighting. Professor Jiang Junjun is currently a tenured professor and doctoral supervisor at the School of Computer Science, Harbin Institute of Technology, vice dean of the School of Artificial Intelligence, and deputy director of the Intelligent Interface and Human-Computer Interaction Research Center. He was selected for the National Youth Talent Program and is also the academic leader of the "Young Scientist Studio" of Harbin Institute of Technology. His research directions involve image processing, computer vision, deep learning (research focuses on large models and image processing, multimodal autonomous unmanned systems, generative artificial intelligence, etc.) and other fields.
Professor Zhang Yongbing is currently a professor and doctoral supervisor at the School of Computer Science at Harbin Institute of Technology. His main research areas include computer vision, biomedical image processing, and computer imaging. In addition, Professor Zhang Yongbing also holds multiple positions. He is a member of many well-known domestic and foreign societies such as the China Computer Society, the China Artificial Intelligence Society, IEEE, SPIE, and OSA. He has published more than 100 papers at international artificial intelligence conferences and has authorized more than 50 invention patents. At present, Professor Zhang Yongbing's main research is to explore the application of artificial intelligence and computer vision in the fields of life medicine and medical health.
In addition to HIT, more and more universities and laboratories are also paying attention to the field of digital pathology and contributing their own strength. For example, a team from Eindhoven University of Technology in the Netherlands published a study titled "A Spatially-Aware Multiple Instance Learning Framework for Digital Pathology", which proposed a model called Global ABMIL (GABMIL). This model is an enhanced version of the traditional ABMIL model. It can integrate spatial information into the embedding vector through the spatial information mixing module, and then use the ABMIL network to predict the slice label, avoiding the traditional MIL method often ignoring a key factor in pathological diagnosis-spatial interaction information between image blocks.
Paper address: https://arxiv.org/abs/2504.17379
In short, the integration of artificial intelligence and traditional medicine is irreversible, and anyone can benefit from it. It is undeniable that it is these "explorers" who are dedicated to the forefront of science that give us the opportunity to enjoy the application of the cross-integration of artificial intelligence and medicine. Of course, with long-term deep cultivation, there is reason to believe that the HIT team will continue to take root here, thereby accelerating the development of the entire field.