Command Palette
Search for a command to run...
Covering More Than 40 Mainstream Models and Data Sets, the Shanghai Jiaotong University Team Released the one-stop Protein Engineering Design Platform VenusFactory
With the rapid development of artificial intelligence computing and data-driven methods, protein engineering is moving towards the AI-assisted design stage. Researchers need more comprehensive, high-quality protein datasets, more powerful and influential protein artificial intelligence models, and more efficient and standardized analysis platforms than ever before, so as to accurately mine valuable information from massive biological data, accelerate the design and optimization of new proteins, and promote innovative breakthroughs in biomedicine, synthetic biology and other fields.
In this context, more and more life science practitioners want to understand AI and use AI technology to assist in the design of protein engineering. However, whether it is David Baker's redesigned open source solution or Meta's ESM series of large models, there are many difficulties in using them, such as the complex logic of the AI computing framework, the large amount of code, and the need for a strong computer programming foundation. In other words, for biological researchers and even non-senior computer practitioners, they still have to face a fairly high threshold for use. In this regard, user-friendly low-code applications have gradually become the mainstream trend in the use of modern open source tools. They can help researchers get rid of complex model configuration and code implementation, allowing computer scientists and biologists to call or train deep learning models in a more convenient way and focus on scientific research itself.
To promote the application and development of artificial intelligence in the field of protein engineering, Professor Hong Liang's research group at Shanghai Jiao Tong University in China developed VenusFactory, a one-stop open platform tailored for protein engineering. Researchers can easily implement tedious data retrieval, model training, task evaluation, model deployment and other functions through interface interaction or command line. Through code-free and process-oriented design, the platform simplifies the complex artificial intelligence engineering operations in the past into lightweight operations at the fingertips, allowing researchers to easily call more than 40 cutting-edge protein deep learning models by starting web services locally without writing complex codes, realizing private data privacy protection, greatly reducing the threshold for intelligent scientific research, and accelerating the in-depth application of artificial intelligence in the field of life sciences.
Code and data are open source at: https://github.com/ai4protein/VenusFactory
Currently, the "VenusFactory Protein Engineering Design Platform" has been launched in the tutorial section of the HyperAI website. The detailed usage tutorial is attached at the end of this article. Interested readers can experience the platform through the link below:
VenusFactory: A unified platform that breaks down barriers to protein AI applications
Protein data is highly dispersed. VenusFactory directly accesses the source of biological data AI protein research is highly dependent on large-scale biological data, and the annotated data is distributed in multiple mainstream public databases. Scientists often need to switch between multiple databases, manually download data, and write scripts for format conversion, resulting in a waste of time and energy on non-practical research work. VenusFactory directly connects to mainstream public databases such as RCSB PDB, UniProt, InterPro, etc. Multi-threaded high-speed download greatly improves the efficiency of data retrieval:
- One-stop access to protein sequence, three-dimensional structure, and functional annotation, fully integrating biological information.
- Standardized format output avoids data compatibility issues and facilitates direct AI training.
- The multi-threaded download mechanism greatly improves the speed of data acquisition, allowing scientists to focus on the research itself.
The evaluation system for protein AI tasks is not unified. VenusFactory covers five core tasks. Currently, the protein AI model evaluation system lacks ready-made authoritative benchmark data, and most research still focuses on the optimization of individual tasks. When researchers choose a solution, they often need to spend a lot of extra time on experimental comparison. VenusFactory integrates more than 40 cutting-edge protein engineering evaluation data sets, covering five core tasks:
- Protein function prediction: Predict the functional tags of proteins to facilitate the discovery of new enzymes and new targets.
- Protein subcellular localization prediction:Predict the localization of proteins in cells to aid disease diagnosis.
- Protein solubility assessment:Improve wet experiment efficiency by pre-judgment of solubility.
- Analysis of the effects of protein mutations: Explore the potential impact of gene mutations and advance precision medicine.
- Other prediction tasks: Such as metal ion binding, protein sorting signal prediction, optimal temperature prediction, etc.
With the help of these benchmark datasets and evaluation results, users can easily compare the performance of different models and select and optimize solutions. At the same time, VenusFactory also provides a download function for all datasets, so users can obtain the corresponding protein sequence, structure, label and other information with one click.
Existing protein AI computational tools have high barriers to use and are difficult for researchers without a computing background to use The use of current protein AI models often requires strong programming skills and deep learning knowledge. For most biologists, training, fine-tuning and applying AI models is still a high-threshold task. VenusFactory integrates more than 40 cutting-edge protein language models (PLMs) in the world, covering comprehensive AI large model solutions, such as the Venus series (ProSST, Pro-Prime, PETA, etc.), ESM series (ESM2, ESM1b, etc.), Ankh series (Base, Large) and ProtTrans series (ProtBert, ProtT5).
- Pre-trained model ecosystem: Directly call open source PLM without training from scratch, saving computing resources.
- High performance fine-tuning: Supports cutting-edge methods such as LoRA and SES-Adapter to adapt the model to specific biological tasks.
- Multitasking support: Whether it is protein solubility prediction or mutant property prediction, you can get started easily.
- Command line mode: Suitable for computer scientists, it can flexibly adjust parameters and achieve deep optimization.
- No-code web interface: Suitable for biologists, you can run AI tasks with simple clicks, no programming knowledge required!
To address these core challenges, VenusFactory has built a one-stop AI-enabled protein engineering platform, providing a complete solution from data acquisition, task evaluation to model fine-tuning, allowing biologists and computational scientists to advance their research efficiently.
Open source & community building to promote scientific innovation
The future of scientific research lies in open sharing. VenusFactory uses the Apache 2.0 license. All codes, data sets, and model weights are completely open source. Users can freely download, modify, and optimize, and share the latest results with researchers around the world. All data, models, and fine-tuning codes are hosted on GitHub & Hugging Face, ensuring that scientists around the world can easily access and reproduce experiments and build their own AI research projects based on VenusFactory.
To help readers experience VenusFactory, HyperAI has launched a one-click deployment tutorial for the "VenusFactory Protein Engineering Design Platform". The following is a detailed introduction to its use.
Tutorial link: https://go.hyper.ai/ZqO3h
VenusFactory Protein Engineering Design Platform Tutorial
Demo Run
1. Log in to hyper.ai, on the Tutorial page, select VenusFactory Protein Engineering Design Platform, and click Run this tutorial online.
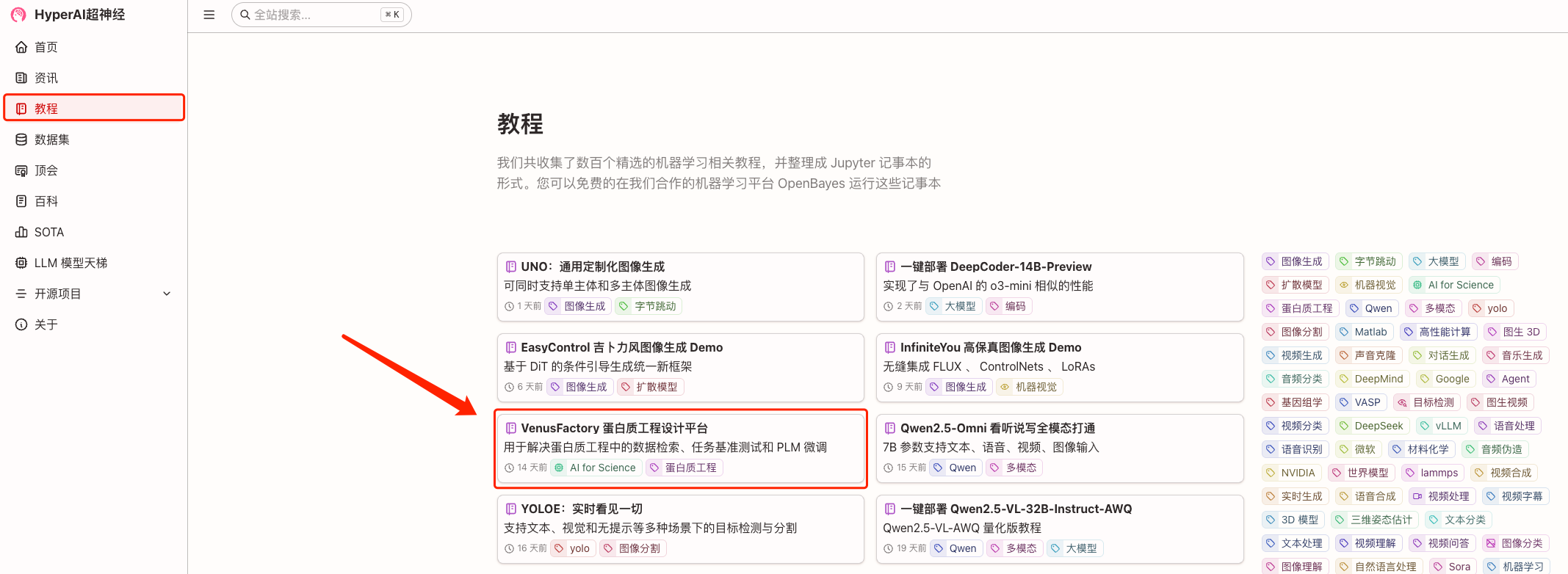
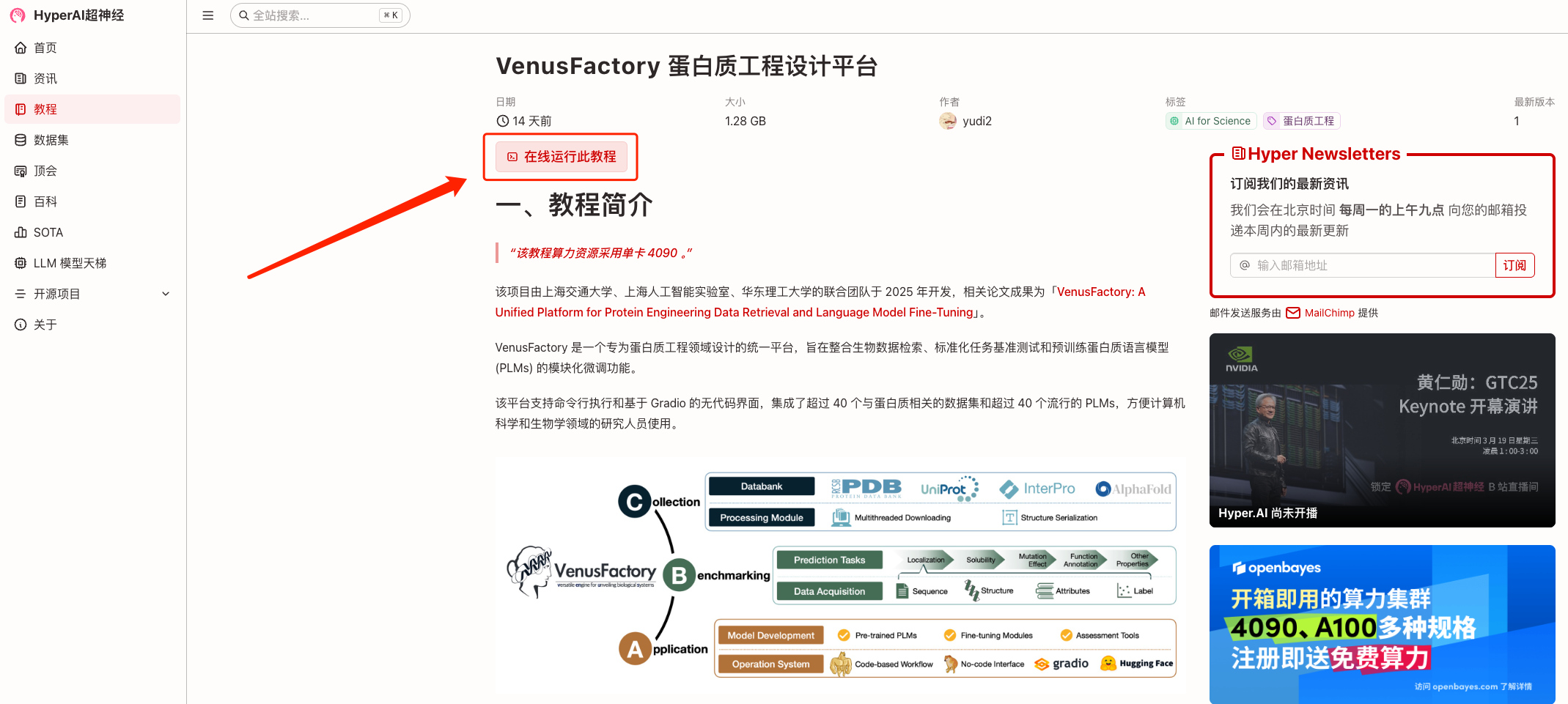
2. After the page jumps, click "Clone" in the upper right corner to clone the tutorial into your own container.
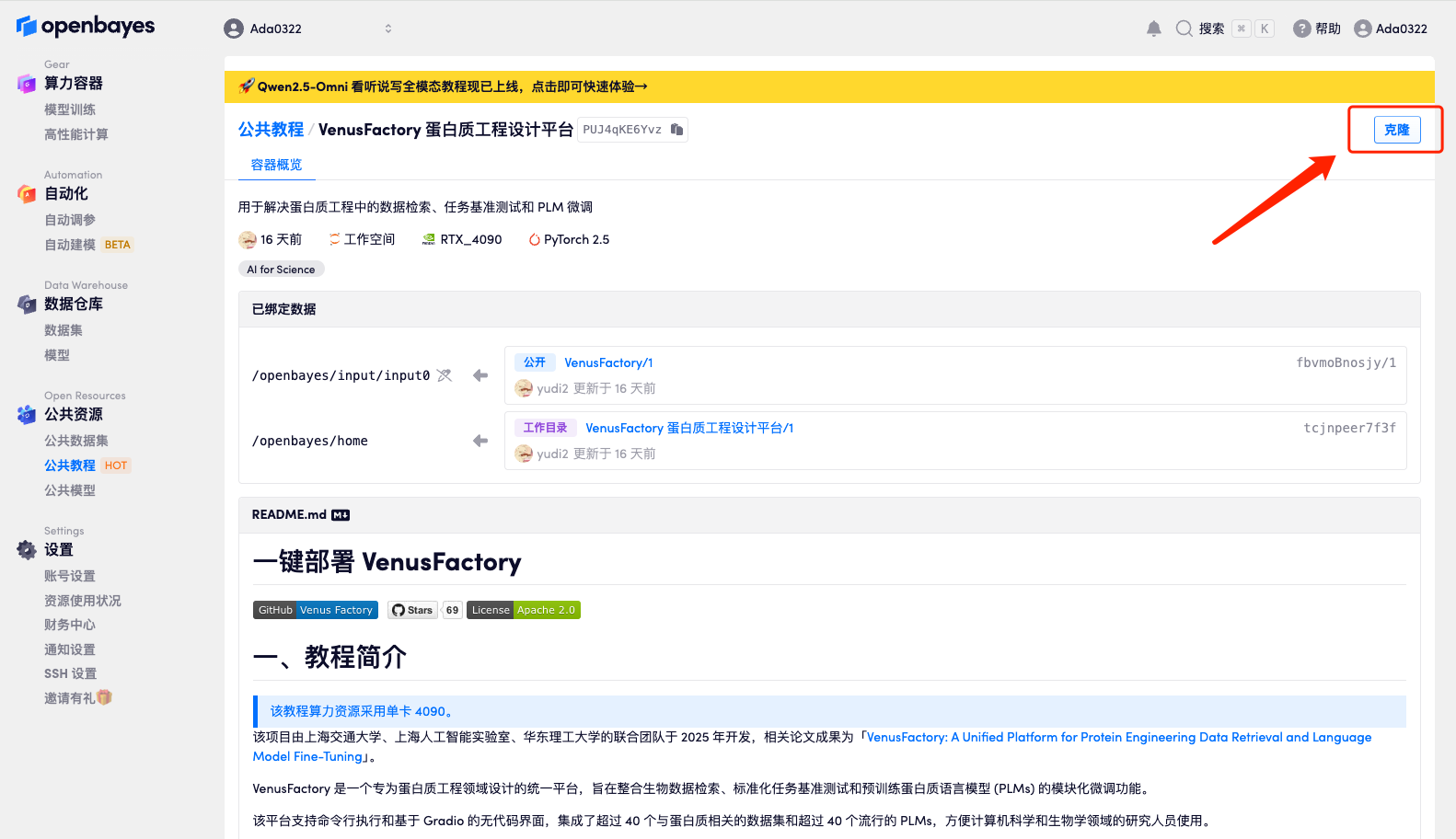
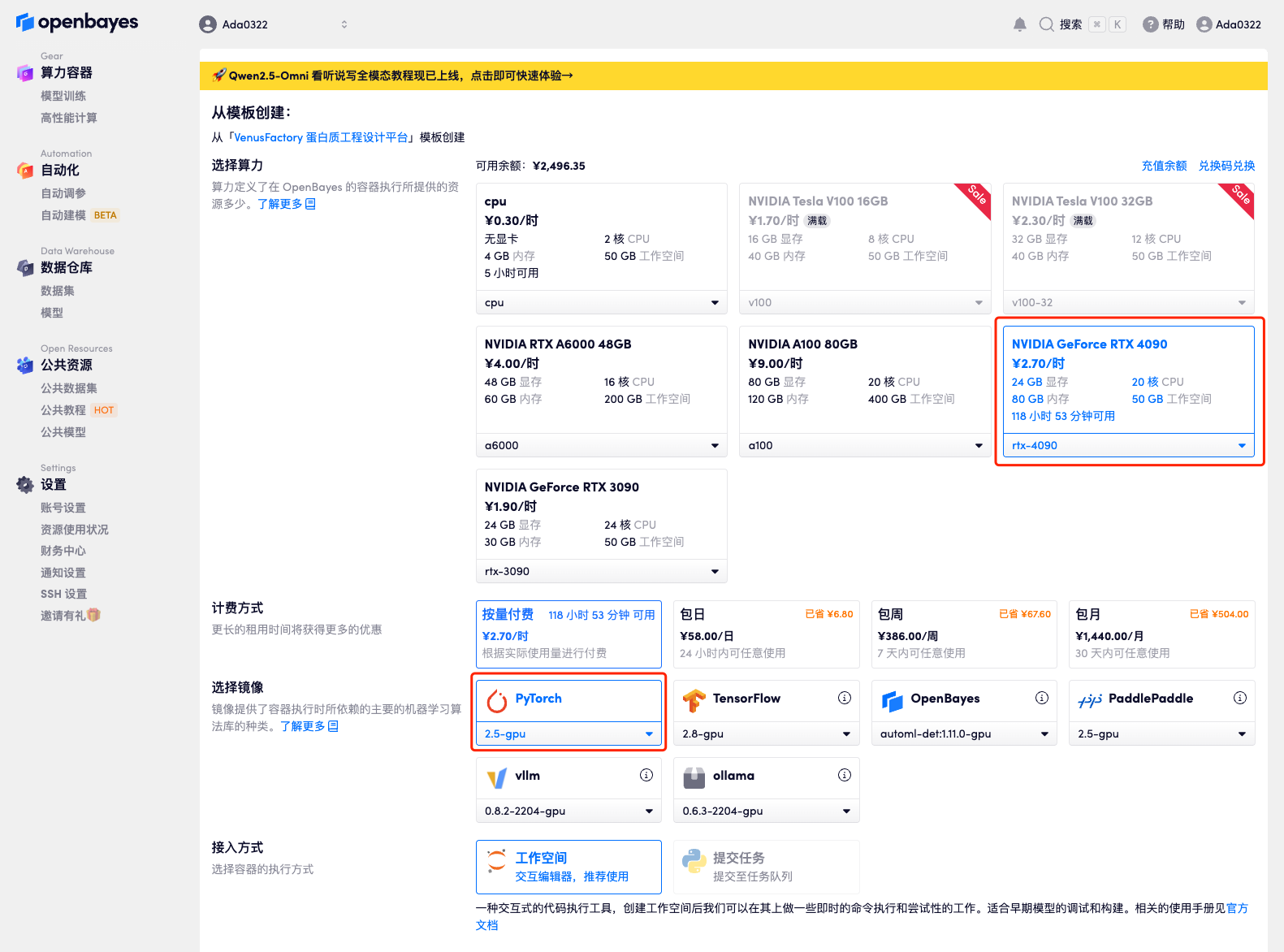
3. Select "NVIDIA GeForce RTX 4090" and "PyTorch" images, and click "Continue". The OpenBayes platform provides 4 billing methods. You can choose "pay as you go" or "daily/weekly/monthly" according to your needs. New users can register using the invitation link below to get 4 hours of RTX 4090 + 5 hours of CPU free time!
HyperAI exclusive invitation link (copy and open in browser):
https://openbayes.com/console/signup?r=Ada0322_NR0n
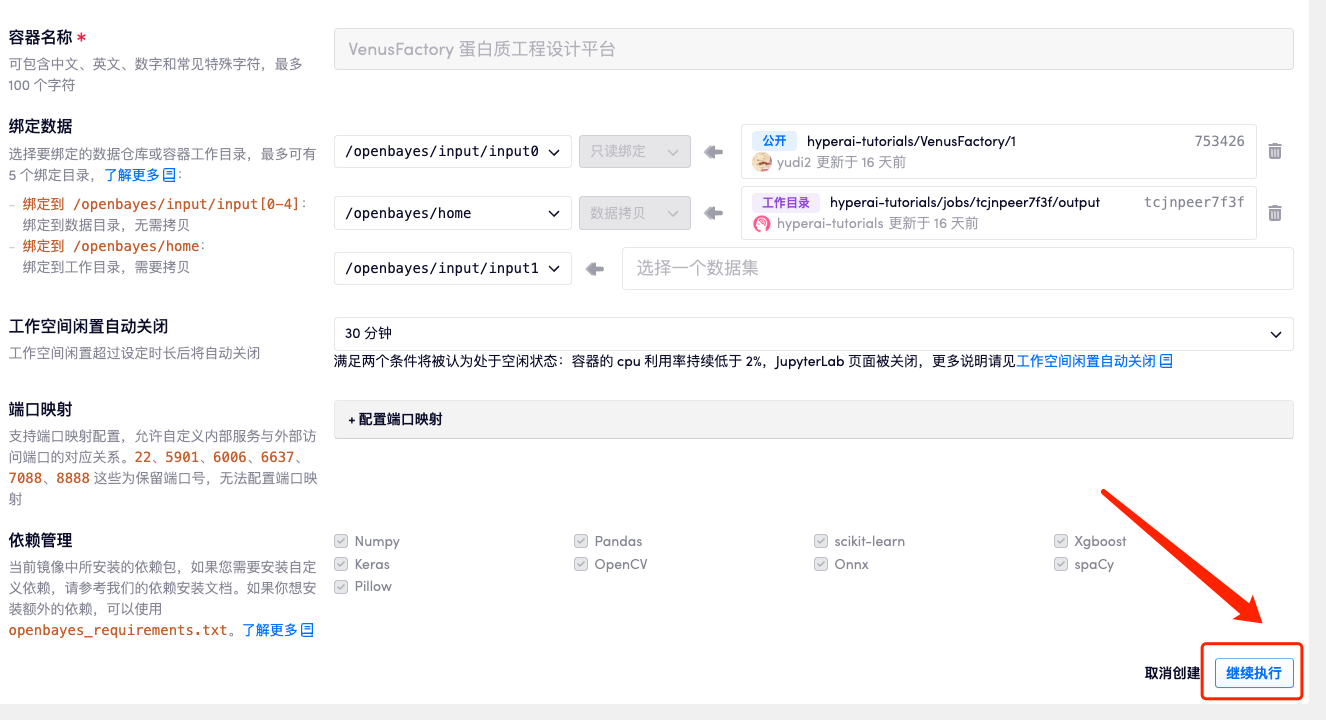
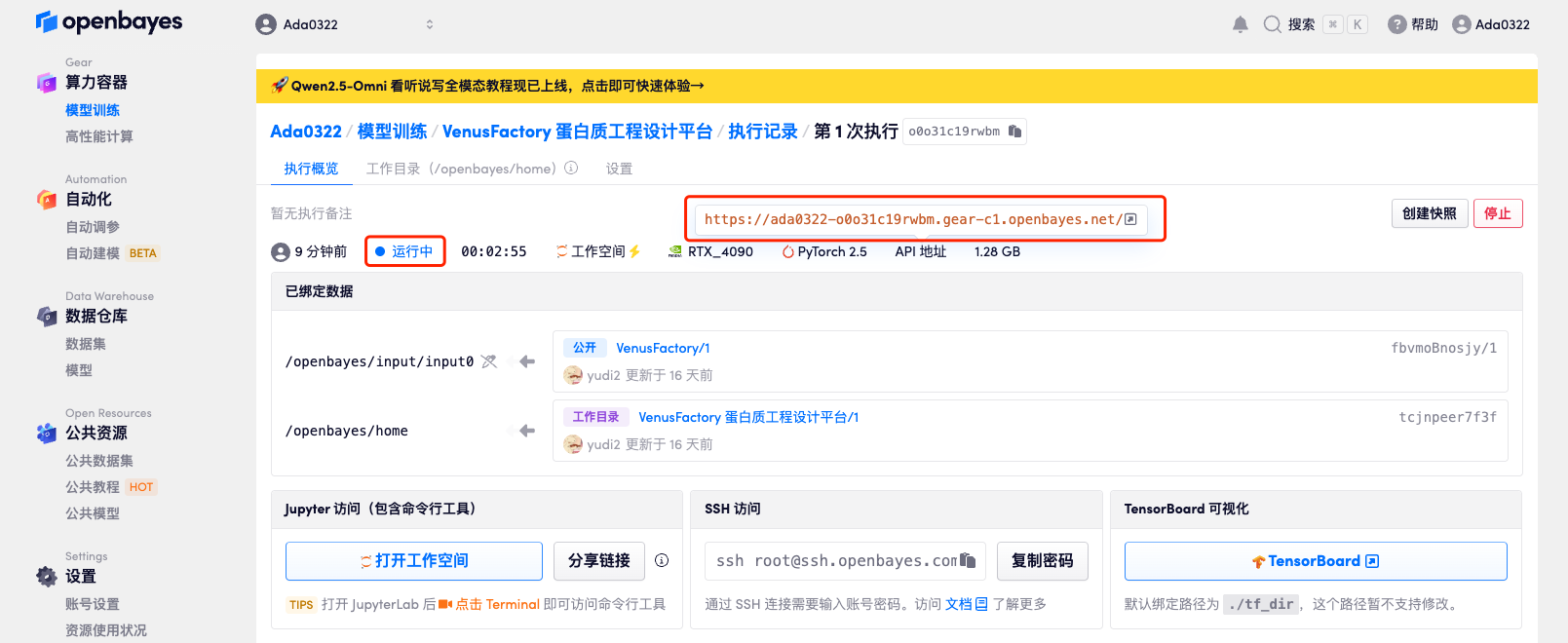
4. Wait for resources to be allocated. The first clone will take about 2 minutes. When the status changes to "Running", click the jump arrow next to "API Address" to jump to the Demo page. Due to the large model, it will take about 3 minutes to display the WebUI interface, otherwise "Bad Gateway" will be displayed. Please note that users must complete real-name authentication before using the API address access function.
Effect display
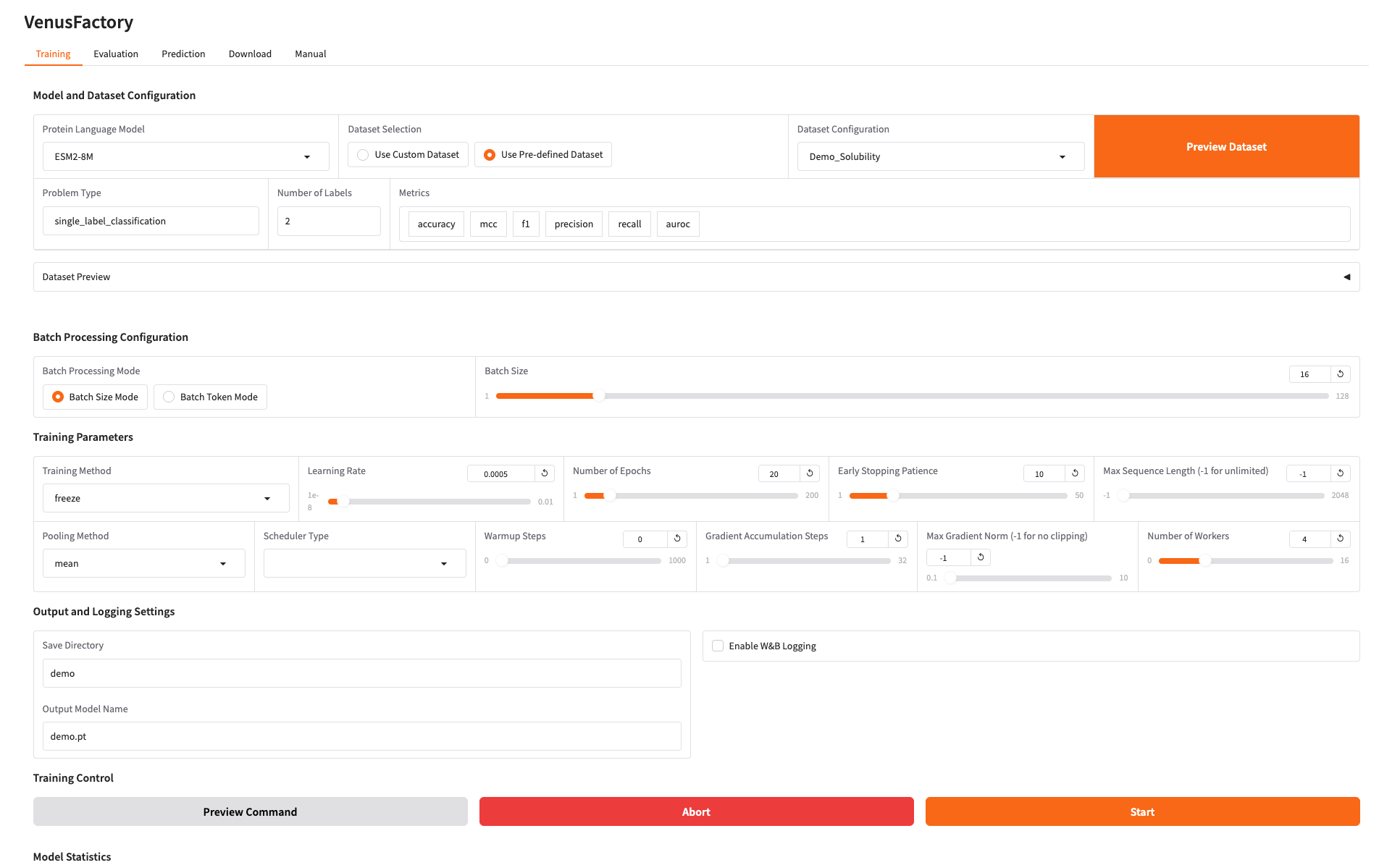
1. This tutorial includes four modules: Training, Evaluation, Predict, and Download. Click Manual and select a language to see detailed instructions for each module.
2. Training module
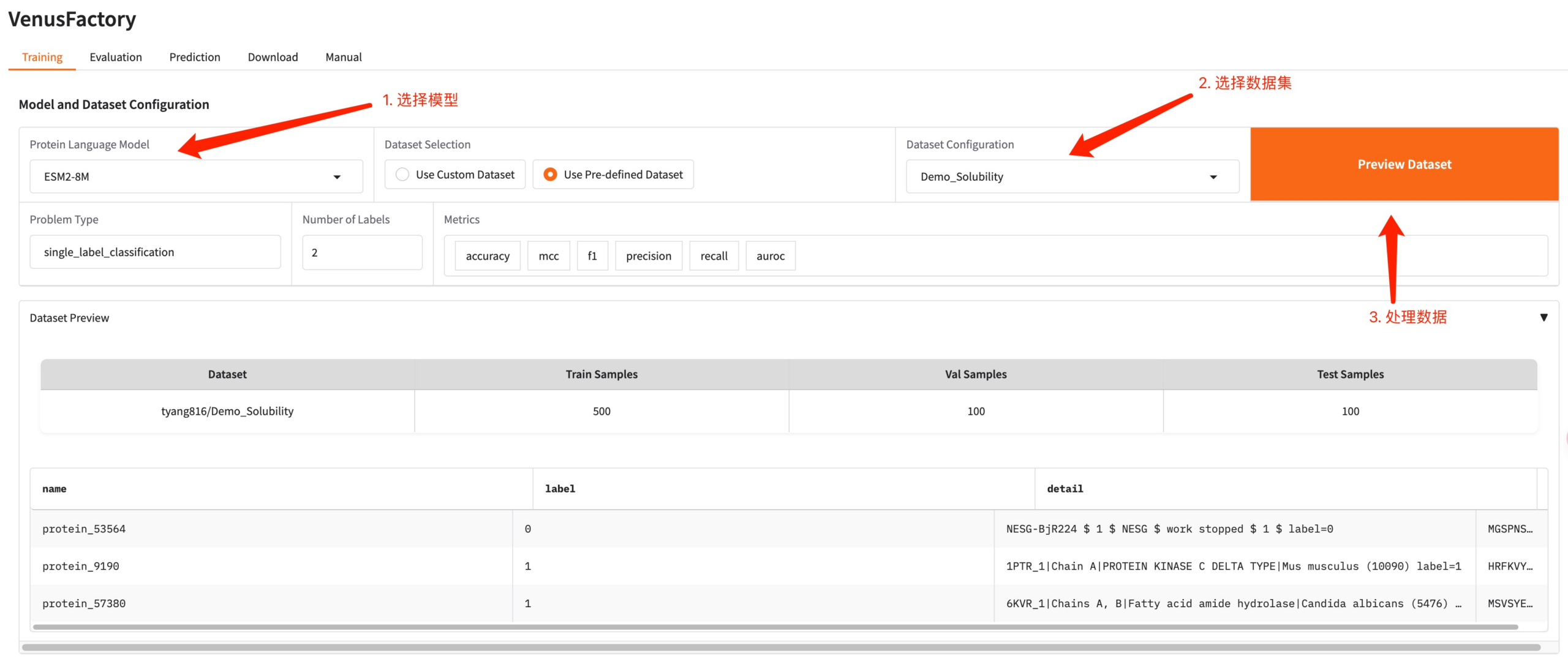
Click the Training module, select the model you want to train in Protein Language Model, and configure the training data in Dataset Configuration
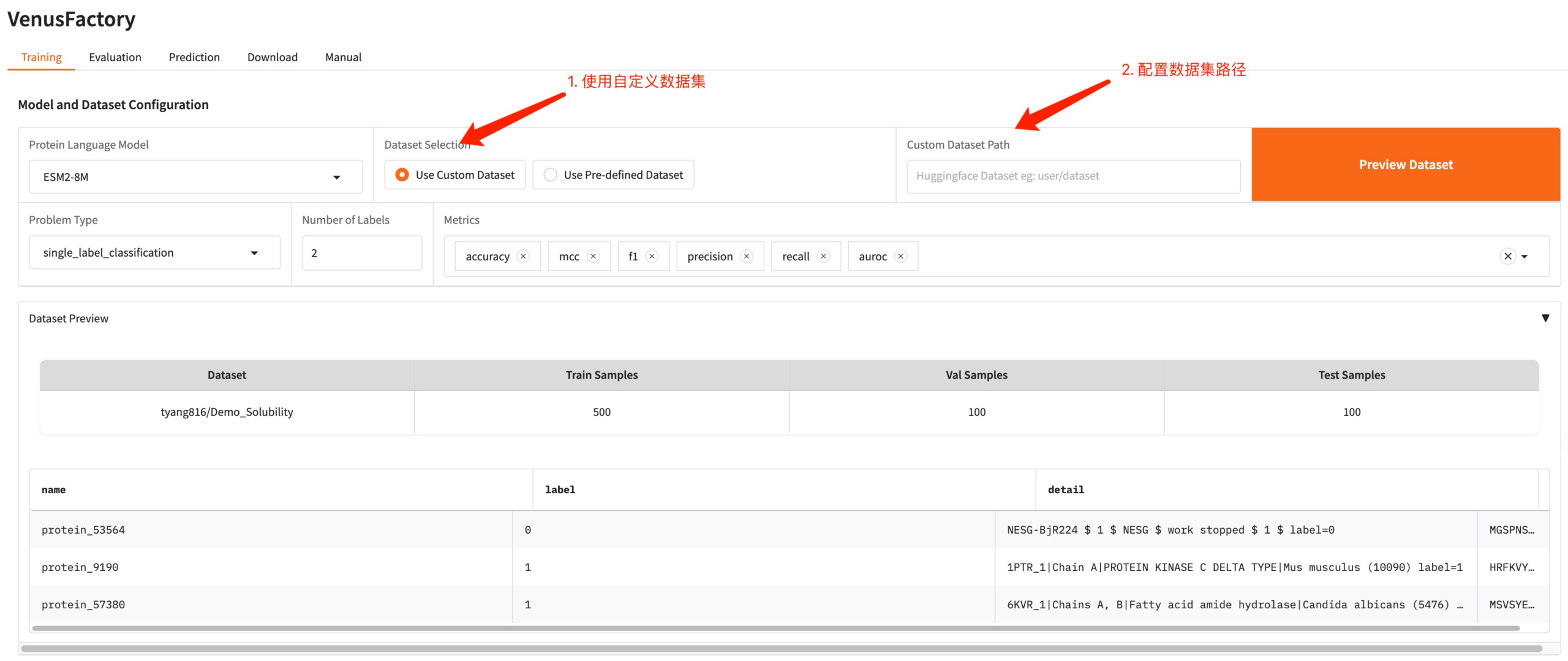
If you need to use your own dataset, you can use the Use Custom Dataset configuration and just need to fill in the dataset path (see the Manual usage documentation for details).
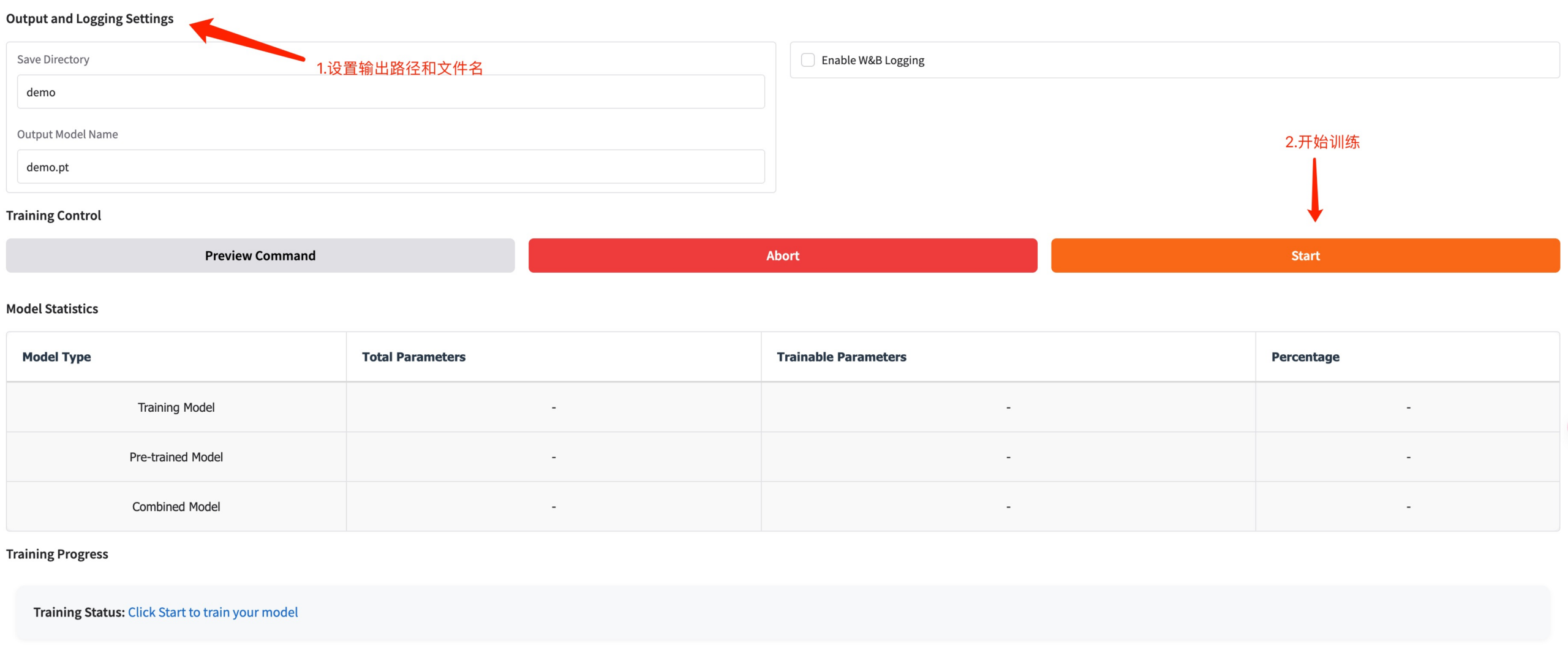
Set the training model save path and click Start to start training.
At this point you can see the training parameters and loss curve
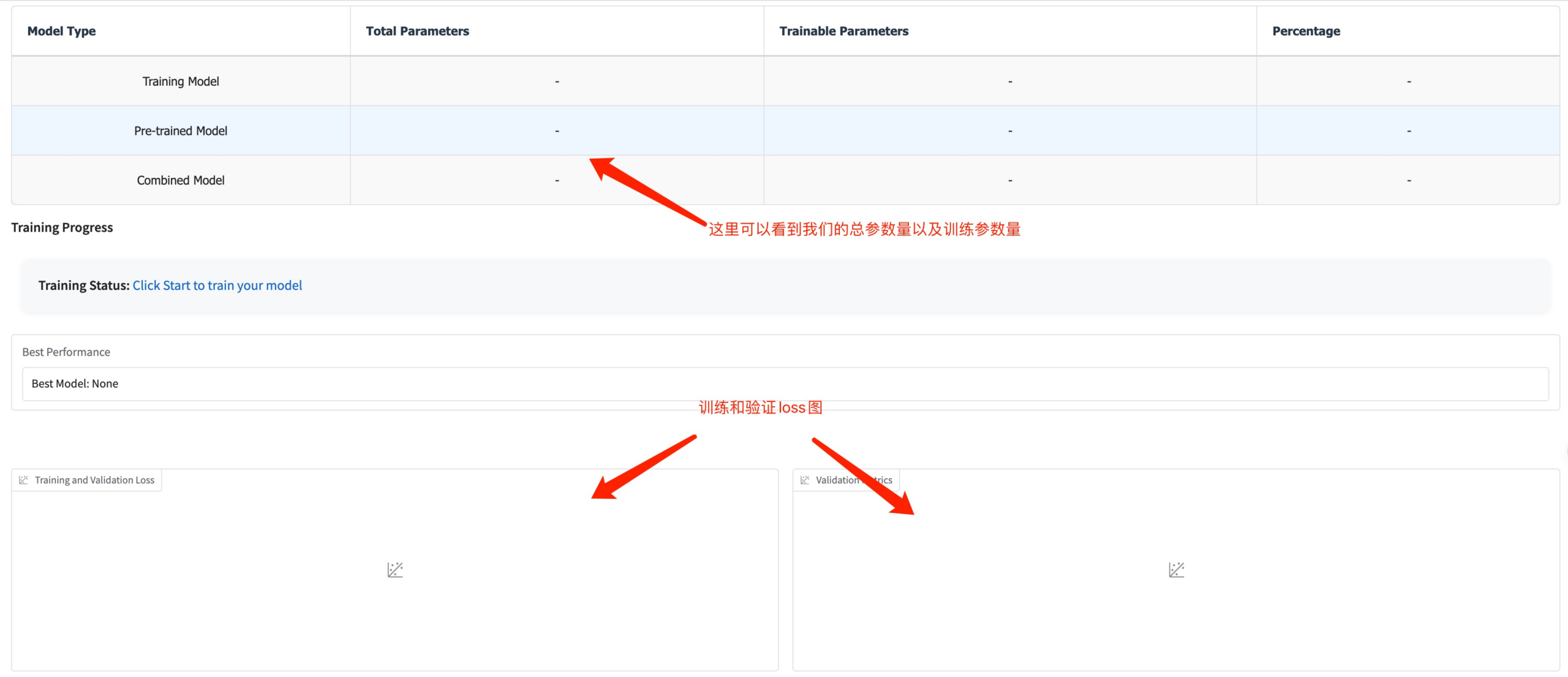
3. Evaluation Module
Click the Evaluation module, configure the model path generated by training and the trained model, process the data, adjust the hyperparameters and start the evaluation.
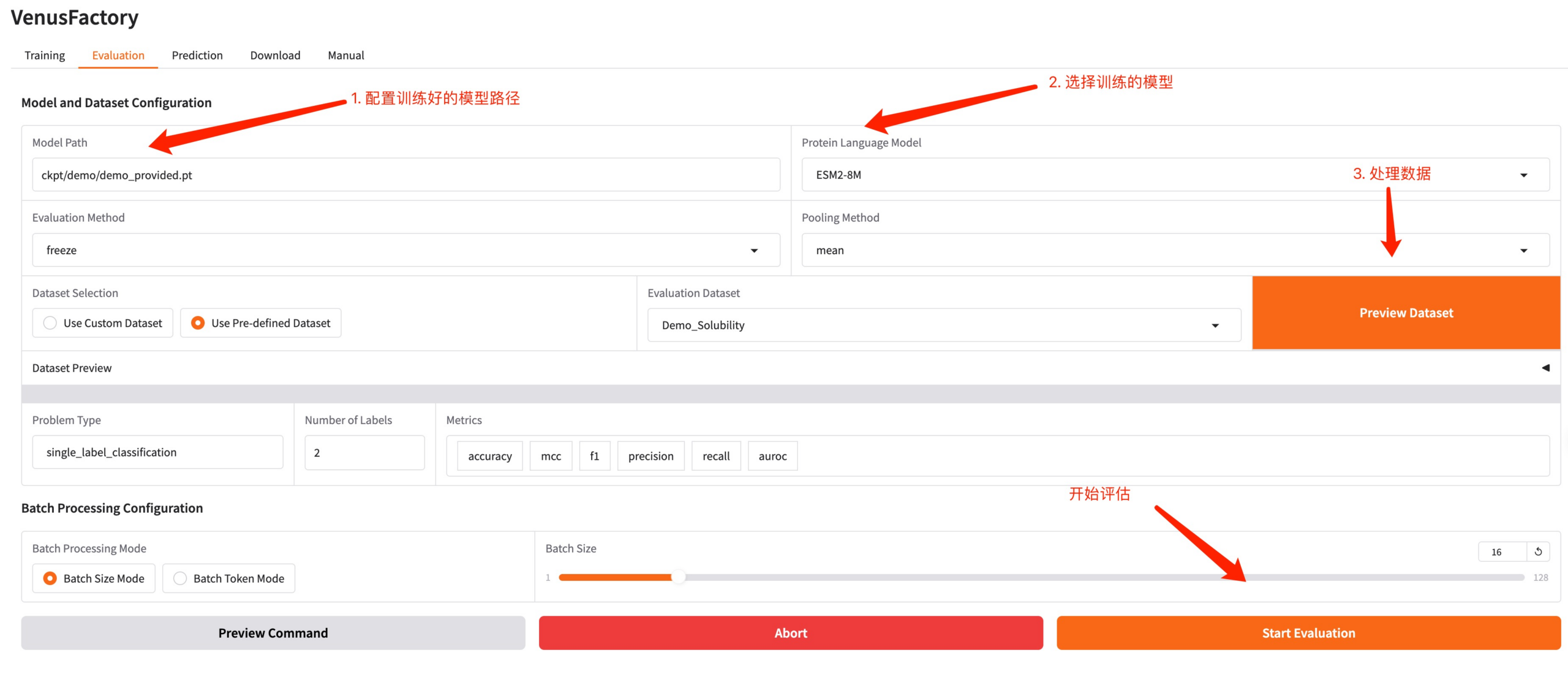
4. Predict Module
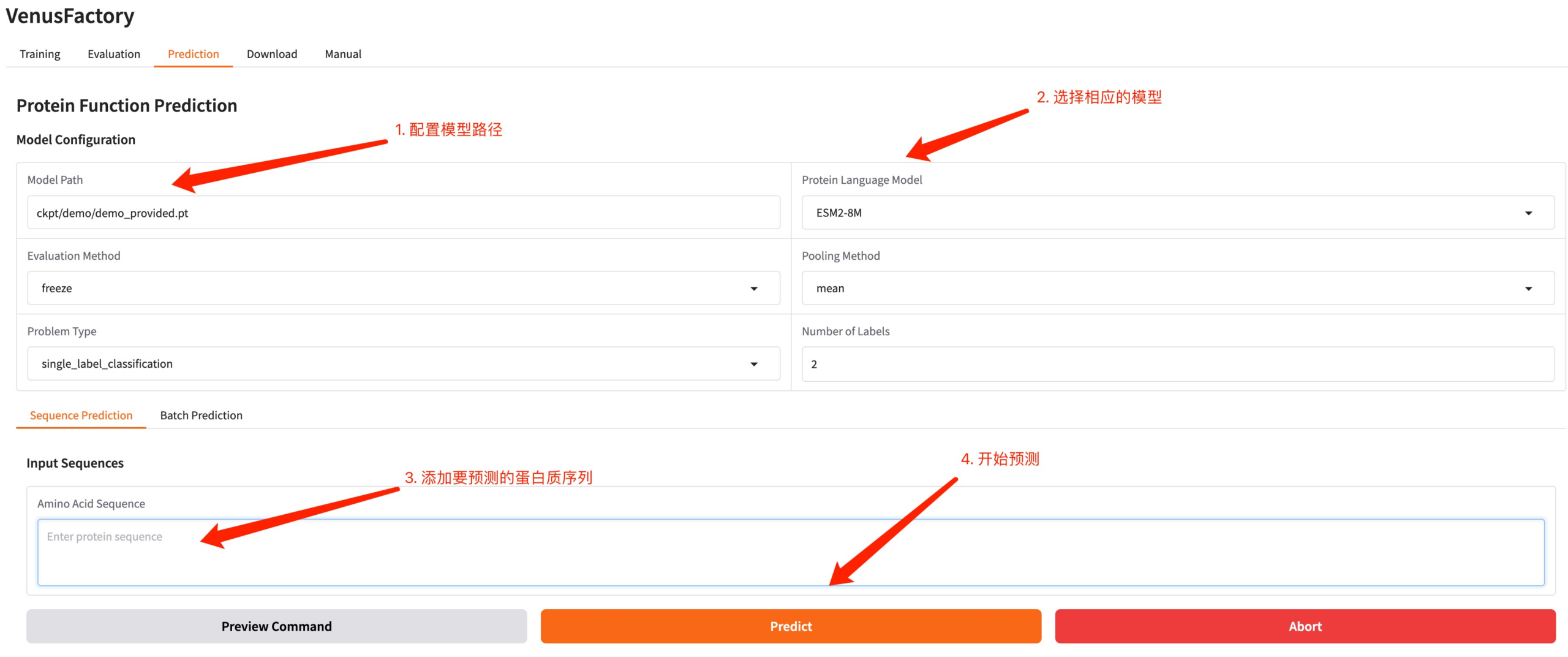
Click the Predict module, configure the model path generated by the training and the trained model, enter the protein sequence you want to predict, and click Predict to make a prediction.
Protein sequence example: MKTWFGHVLQ
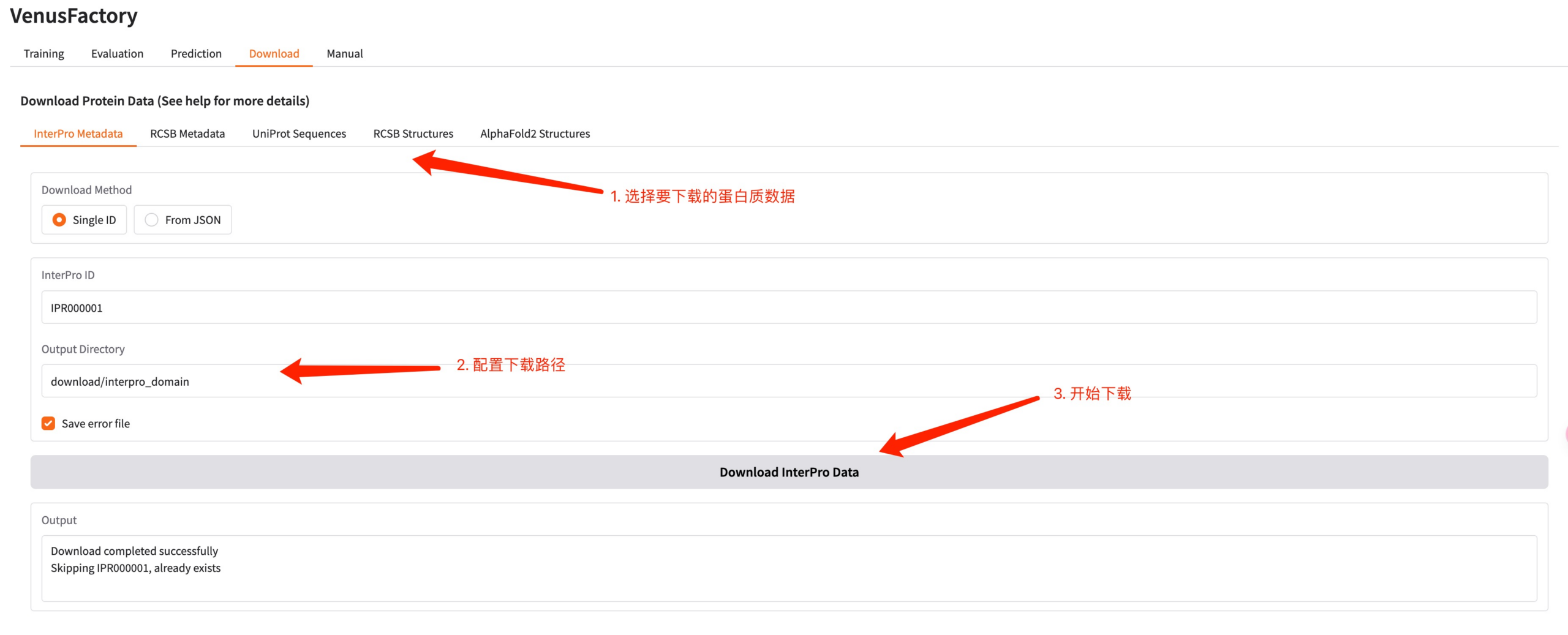
5. Download module
Click the Download module to download protein data in this interface.
The above is a detailed tutorial on how to use the "VenusFactory Protein Engineering Design Platform". Everyone is welcome to come and experience it!








