Command Palette
Search for a command to run...
For the First Time, the Tsinghua Team Has Achieved the Unification of Molecular Generation and Property prediction. It Has Proposed a two-stage Diffusion Generation Mechanism and Was Selected for ICLR 2025.

Artificial intelligence technology is profoundly reshaping the process of drug development.Among them, molecular property prediction and molecular generation, as two core tasks, have long developed along independent technical paths.The purpose of molecular property prediction is to predict the diverse chemical and biological properties of molecules given molecular structure information, and to accelerate drug screening. Molecular generation aims to estimate the distribution of molecular data, potentially learn atomic interactions and conformational information, and be able to generate new chemically reasonable molecules from scratch, thus expanding the boundaries of drug design possibilities. Although a lot of research has been done in these areas in recent years, they have basically developed independently.It has never been possible to effectively open up the collaborative channels between these two key links.
In view of this,Tsinghua University and the Chinese Academy of Sciences team proposed the UniGEM model, which for the first time achieved collaborative enhancement of two tasks based on a diffusion model.The research team pointed out that generation and property prediction are highly correlated and rely on effective molecular representation. The team innovatively proposed a two-stage generation process, overcoming the inconsistency in traditional joint training and opening up a new path in the field of molecular generation and property prediction. The achievement was selected for ICLR 2025 under the title "UniGEM: A Unified Approach to Generation and Property Prediction for Molecules".
Paper address:
https://openreview.net/pdf?id=Lb91pXwZMR
QM9 quantum chemistry dataset:
GEOM-Drugs 3D molecular conformation dataset:
The open source project "awesome-ai4s" brings together more than 200 AI4S paper interpretations and provides massive data sets and tools:
https://github.com/hyperai/awesome-ai4s
Motivation for unifying generation and prediction tasks
The research team believes that the essence of both generation and prediction tasks lies in learning molecular representations.On the one hand, the effectiveness of various molecular pre-training methods shows that molecular property prediction relies on robust molecular representation as a foundation. On the other hand, molecular generation requires a deep understanding of molecular structure to be able to create good representations during the generation process.
Recent research results provide support for this view. For example, work in the field of computer vision has shown that diffusion models have the ability to learn effective image representations. In the molecular field, studies have shown that generative pre-training can enhance molecular property prediction tasks, but these methods usually require additional fine-tuning to achieve optimal prediction performance. In addition, although predictors can guide molecular generation through classifier guidance methods, it is still unclear whether training predictors can directly improve generation performance.
Therefore, existing research has not yet fully clarified the relationship between generation tasks and prediction tasks.This raises a key question: Can we build a unified model that achieves synergistic enhancement of generation and prediction tasks?
Analysis of the reasons for failure of traditional methods
A straightforward approach to combining these two tasks is to use a traditional multi-task learning framework, where the model optimizes both the generation loss and the prediction loss. However, the research team's experiments showed that this approach significantly degraded the performance of both the generation task and the property prediction task (generation stability dropped by 6%, and the prediction error increased by more than 1 times simultaneously). Even when freezing the weights of the generation model and adding a separate head for the property prediction task to maintain generation performance, the researchers observed no improvement in property prediction performance compared to training from scratch.
The researchers attribute the poor results of traditional methods to the inherent inconsistency between generation and prediction tasks.During the diffusion generation process, the molecular structure needs to undergo a gradual reconstruction from disordered noise to fine structure. However, in the prediction task, meaningful molecular properties can only be defined after the molecular structure is basically established. Therefore, simply using a simple multi-task optimization method will result in highly disordered molecular conformations being incorrectly associated with property labels in the early diffusion stage, which will have a negative impact on molecular generation and property prediction.
To further illustrate this point, the researchers conducted a theoretical analysis of the mutual information between the intermediate representations within the denoising network and the target molecules during diffusion training.We then theoretically prove that the diffusion model implicitly maximizes the lower bound of the mutual information between the intermediate representation and the target molecule, indicating the ability of the diffusion model representation learning. However, the mutual information between the intermediate representation and the target molecule shows a monotonically decreasing trend and approaches zero at larger time steps, which means that the intermediate representation in the disordered stage cannot support effective prediction. Therefore, both intuition and theory show that the generation task and the prediction task can only be aligned at smaller time steps, that is, when the molecules remain relatively ordered.
Two-stage diffusion generation mechanism
Based on the above analysis,The research team proposed a novel two-stage generation method that aims to unify the prediction and generation of molecular properties, as shown in the figure below.
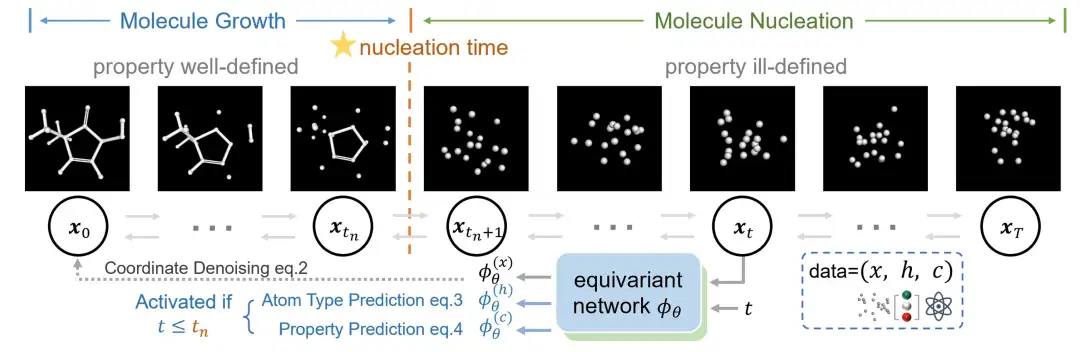
The researchers divided the molecule generation process into two stages, namely the "molecular nucleation stage" and the "molecular growth stage",This division is inspired by the crystal formation process in physics.
During the molecular nucleation stage, the molecule forms its skeleton from a completely disordered state, and then a complete molecule grows based on the skeleton. These two stages are separated by the "nucleation time". The researchers introduced a new way of molecular generation to describe these two stages. Among them, before the "nucleation time", the diffusion model gradually generates molecular coordinates; after nucleation, the model continues to adjust the molecular coordinates while optimizing the properties and atomic type prediction losses.
Different from traditional generative models that usually perform joint diffusion of atom types and coordinates, this innovative method only focuses on the diffusion of coordinates and treats atom types as a separate prediction task.Because the researchers observed that the atom types can often be inferred from the formed molecular coordinates. Specifically, before nucleation, the diffusion process aims to reconstruct the coordinates; after nucleation, it integrates the prediction loss of atom types and properties into a unified learning framework.
UniGEM training strategy
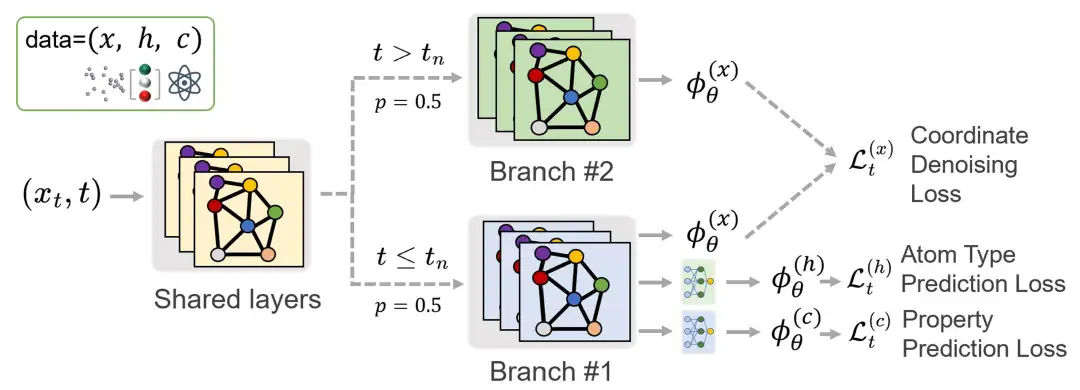
In order to facilitate comparison with the traditional joint diffusion method, the researchers used the E(3) equivariant diffusion model (EDM) using EGNN as the network structure skeleton. Among them, the growth phase only accounts for about 1% of the entire training process. If the standard diffusion training procedure is followed and the time step is uniformly sampled, the number of iterations of the prediction task only accounts for 1% of the total training process, which will significantly reduce the performance of the model on this task.Therefore, to ensure adequate training for the prediction task, the researchers oversampled the time steps during the growing phase.
However, the researchers observed that oversampling can lead to imbalanced training across the entire time step range, which in turn affects the quality of the generation process. To address this problem, they proposed a multi-branch network architecture. The network shares parameters at shallow levels, but splits into two branches at deeper levels, each with an independent set of parameters.These branches are activated at different stages of training: one branch focuses on the nucleation phase, another handles the growth phase,As shown in the figure below, this design ensures that the prediction task and the generation task can be trained effectively without affecting each other.
UniGEM’s reasoning process
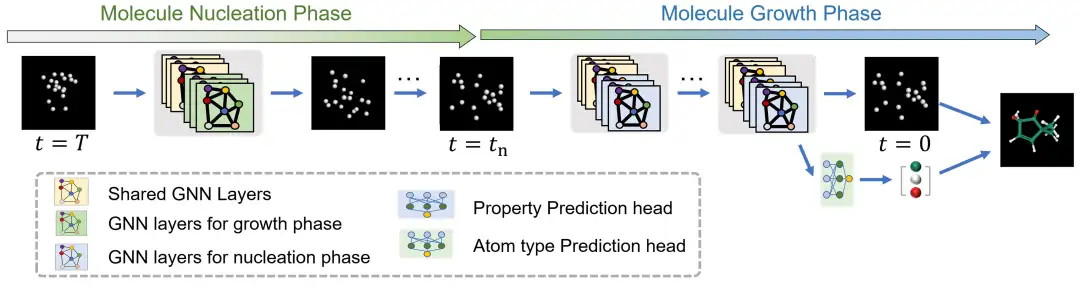
In UniGEM,Molecular generation is done by reconstructing atomic coordinates through a back-diffusion process and then predicting the atomic type based on the generated coordinates.As shown in the figure. For property prediction, the time step of the fixed network input is zero, and the property prediction head is used. It is worth noting that this method does not incur additional computational overhead for both the generation task and the prediction task, and the total inference time is the same as the baseline.
For the molecular generation task, the researchers also analyzed the differences in generation errors between UniGEM and traditional joint generation methods.First, it is observed that in UniGEM, the error of the atom type prediction loss is smaller than the atom type denoising generation loss in joint generation. Second, during the joint generation process, the coordinate generation is affected by the oscillation of the atom type prediction results, which leads to an increase in error. Finally, the joint generation method also introduces larger initial distribution errors and discretization errors. These factors together explain how UniGEM achieves better generation results.
Experimental results: Outperform the baseline model in both molecular generation and property prediction tasks
Molecular Generation: UniGEM Outperforms Benchmark Models
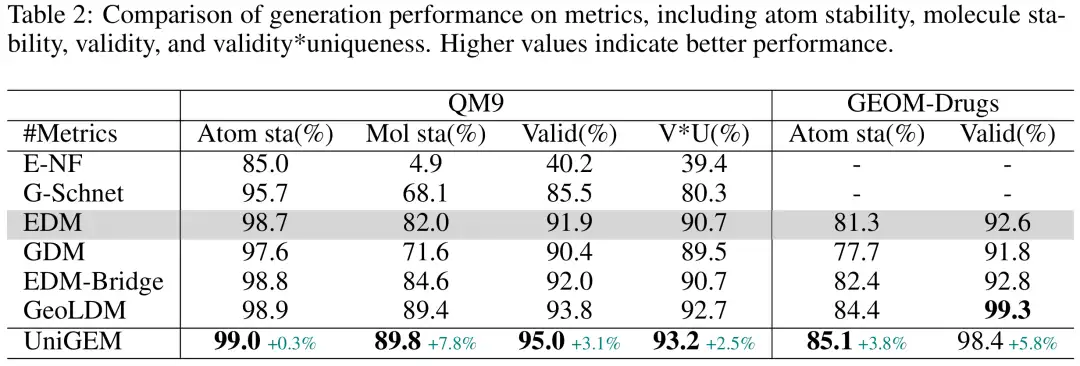
The researchers first compared UniGEM based on EDM with EDM variants on the QM9 and GEOM-Drugs datasets. UniGEM outperformed the baseline model in almost all evaluation indicators, as shown in the figure below. It is worth noting that compared with other EDM variants,UniGEM is significantly simpler as it neither relies on prior knowledge nor requires additional autoencoder training, yet it outperforms EDM-Bridge and GeoLDM, highlighting the advantages of UniGEM.
To demonstrate the flexibility of UniGEM in adapting to various generation algorithms, the researchers applied UniGEM to Bayesian flow networks (BFNs), surpassing GeoBFN, which jointly generated coordinates and atom types, on the QM9 dataset, achieving SOTA results.
In addition, the researchers tested UniGEM’s performance in conditional generation tasks, avoiding the need to retrain the conditional generation model by using the model’s own property prediction module as a guide during the sampling process.
Molecular property prediction: UniGEM surpasses most pre-training methods
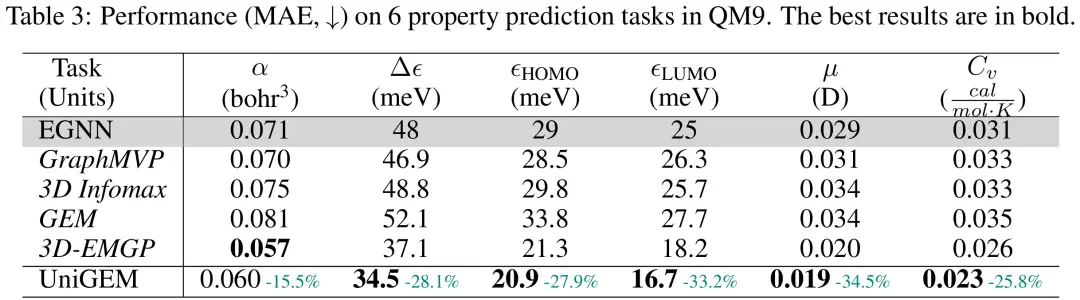
The researchers evaluated the performance of UniGEM property prediction on the QM9 dataset, using the mean absolute error (MAE) on the test set as the evaluation metric.UniGEM significantly outperforms EGNN trained from scratch, demonstrating the effectiveness of unified modeling.Surprisingly, despite these state-of-the-art pre-training methods leveraging additional large-scale pre-training datasets, UniGEM still outperforms most pre-training methods. This highlights the advantage of its unified model for generation and prediction, which can effectively leverage the power of molecular representation learning in the generation process without the need for additional data and pre-training steps.
Conclusion
The UniGEM model unifies the tasks of molecule generation and property prediction and significantly improves the performance of both. The enhanced performance of UniGEM is supported by solid theoretical analysis and comprehensive experimental studies. We believe that the innovative two-stage generation process and its corresponding model provide a new paradigm for the development of molecular generation frameworks and may inspire the development of more advanced molecular generation frameworks, thereby benefiting molecular generation in more specific application fields.
This research is led by ATOM Lab. The team has more research results in the fields of molecular pre-training, molecular generation, protein structure prediction, virtual screening, etc., so please pay attention!
Welcome to ATOM Lab homepage:
https://atomlab.yanyanlan.com/
About the author:
* Lan Yanyan is a professor at the Institute of Intelligent Industries (AIR) at Tsinghua University. Her research interests include AI4Science, machine learning, and natural language processing.
* Shikun Feng is a PhD student at the Institute of Intelligent Industries (AIR) at Tsinghua University. His research interests include representation learning, generative models, and AI4Science.
* Yuyan Ni is a PhD candidate at the Academy of Mathematics and Systems Science (AMSS) of the Chinese Academy of Sciences. Her research interests include generative models, representation learning, AI4Science, and deep learning theory.
The main authors of this paper, Dr. Shikun Feng and Dr. Yuyan Ni, are currently looking for job opportunities. Interested friends can contact them.
* Feng Shikun's email: [email protected]
* Ni Yuyan's email address: [email protected]