Command Palette
Search for a command to run...
Oxford/Amazon/Westlake University/Tencent and Others Proposed a Multimodal, Multi-domain, multi-language Medical Model M³FM, Which Can Be Used for zero-sample Clinical Diagnosis

I believe that many friends who love Marvel movies have been amazed by this scene. In the movie "Iron Man 2", the artificial intelligence butler Jarvis collected Stark's blood samples and used deep learning algorithms to quickly modularize the sample data. It accurately and quickly analyzed the palladium content in Stark's body, and even gave cross-domain suggestions while issuing a report, such as "existing elements cannot replace palladium metal, and new elements need to be synthesized."Although this is only a few tens of seconds of footage, it perfectly demonstrates the automation, intelligence, and process-based features of smart healthcare.
However, in real life, in order to achieve the same result, medical staff need to go through complicated processes such as blood sampling, image analysis, data comparison, report issuance, and disease classification. And this is only from a macro perspective, and it is even worse if it is subdivided. Taking the most common medical images in clinical diagnosis as an example, medical images can describe clinical findings and provide a basis for further diagnosis of diseases. But when it comes to accurately, concisely, completely, and coherently describing a report on medical images in natural language, it will make many medical staff feel headache and boring.Statistics show that even for experienced doctors, it usually takes an average of 5 minutes or more to complete a report.
Fortunately, although science fiction has not yet fully illuminated reality, it has already revealed a glimmer of light through the cracks in the darkness. In the intersection of artificial intelligence and medical health, more and more researchers have conducted extensive research and developed methods for automatic report generation. These methods can automatically generate draft reports for medical staff to review, modify and refer to. On the one hand, they can effectively solve the time-consuming and labor-intensive work tasks of medical staff, and on the other hand, they can reduce the probability of human errors through automation.
Recently, npj Digital Medicine, a journal under the internationally renowned academic journal Nature Portfolio, published a study titled "A multimodal multidomain multilingual medical foundation model for zero shot clinical diagnosis".It mentions a multimodal (image and text), multidomain (CT and CXR), and multilingual (Chinese and English) medical foundation model M³FM (Multimodal Multidomain Multilingual Foundation Model), which can be used for zero-sample clinical diagnosis and support disease reporting and disease classification.The researchers demonstrated the effectiveness of this method on nine benchmark datasets for two infectious and 14 non-communicable diseases, outperforming previous methods.
The study has a luxurious lineup of authors. In addition to teams from Oxford University, University of Rochester, Amazon and other institutions, it also includes Dr. Zheng Yefeng from the Medical Artificial Intelligence Laboratory of Westlake University and Dr. Wu Xian, head of the Tianyan Research Center of Tencent Youtu Lab.

Paper address:
https://www.nature.com/articles/s41746-024-01339-7
The open source project "awesome-ai4s" brings together more than 200 AI4S paper interpretations and provides massive data sets and tools:
https://github.com/hyperai/awesome-ai4s
Data loss is still a pain point for existing methods
Medical imaging is the basis of medical imaging reports and disease classification, and plays an important role in subsequent clinical diagnosis. Therefore, the research on related automation methods has naturally become one of the focuses of scientific research. However, despite the fruitful research results, there are still many deficiencies from a practical perspective.Among them, the scarcity or even complete lack of data is a key challenge.
on the one hand,The generation of disease reports is similar to the image-based language generation task, which aims to generate descriptive text to describe the input image. Traditional basic methods usually rely heavily on a large amount of high-quality medical training data annotated by clinicians, which is expensive and time-consuming to collect, especially for rare diseases and non-English languages.
Specifically, for new or rare diseases, these diseases usually lack sufficient valid data for training in the early stages. For example, the COVID-19 pandemic that began to rage around the world at the end of 2019 had limited data available in the early stages, which resulted in system training taking much longer than the duration of the first few waves of the pandemic. According to the "2024 China Rare Disease Industry Trend Observation Report", there are more than 7,000 known rare diseases in the world. Conservative evidence-based data estimates that the prevalence of rare diseases in the population is about 3.5% to 5.9%, and the number of people affected by rare diseases worldwide is approximately 260 million to 450 million. Such a large but atypical disease undoubtedly makes the above problems more challenging.
In addition, the global healthcare system involves different regions, different populations, and different languages. For languages other than English, the relevant labeled data is usually very scarce or even completely missing. Therefore, the limited labeled data undoubtedly poses a major challenge to the non-English language training system using existing methods. At the same time, this also makes it more difficult for existing methods to deal with uncommon languages, further affecting the goal of AI fairness and failing to fully benefit underrepresented groups.
on the other hand,In order to effectively classify diseases, the current advanced models are mostly inspired by the success of CLIP, such as BioViL, REFERS, MedKLIP, and MRM, which are all developed to better understand medical multimodal data. In the implementation process, these methods use contrastive learning to pre-train the CLIP model using medical data, but since most models are specific to chest X-ray (CXR), they are generally unable to process multi-domain, multi-language medical images and texts within a single framework. At the same time, previous work has also been unable to perform zero-sample disease reporting for different fields of language and images.
* The CLIP model is a contrastive language-image pre-training model developed by OpenAI - an effective method for learning from natural language supervision. CLIP mainly learns the association between images and text through contrastive learning, and pre-trains on large-scale image-text pairs, so that the model can understand and associate information from different modalities.
In this context, it is urgent to develop a model that can perform multimodal, multi-domain, and multi-language clinical diagnosis with few or zero samples.The specific innovations proposed in this study are as follows:
* The proposed M³FM is the first attempt to conduct zero-shot multimodal multi-domain multi-language clinical diagnosis, where labeled data for training is scarce or even completely missing;
* M³FM validates its effectiveness on 9 datasets, including two domains of medical imaging data, namely CXR and CT; two different languages, namely Chinese and English; two clinical diagnosis tasks, namely disease reporting and disease classification; and multiple diseases, including 2 infectious and 14 non-infectious diseases.
M³FM: Two major modules, verified by multiple data sets
In this study, the key idea of the proposed M³FM is to pre-train the model on public medical data across modalities, domains, and languages in order to learn extensive knowledge and then leverage this knowledge to accomplish downstream tasks without the need for labeled data. The main components of the M³FM framework include 2 main modules,That is, MultiMedCLIP and MultiMedLM. As shown in the figure below:
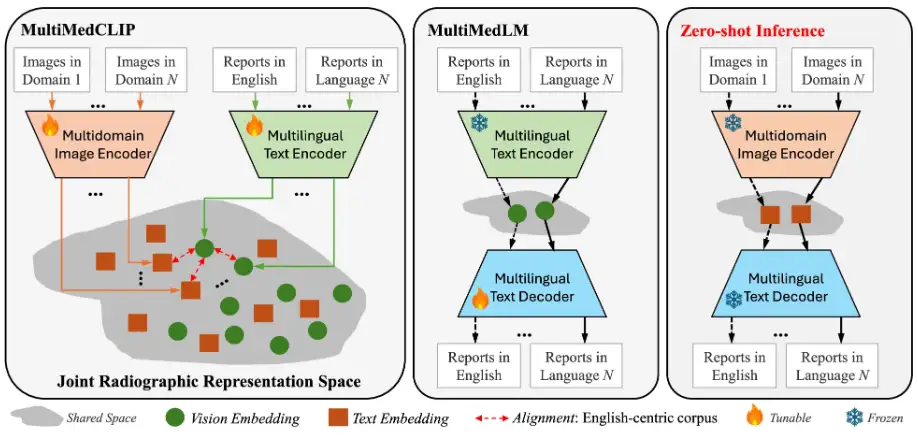
The process is that MultiMedCLIP aligns and bridges different languages and images in a shared common latent space.MultiMedLM then reconstructs the text based on the text representation in a shared latent space, and finally M³FM generates multilingual reports directly based on the visual representations of input images from different domains in a unified latent space.
Specifically, MultiMedCLIP is a module for learning joint representations. It introduces a multi-domain visual encoder and a multi-language text encoder, with the goal of creating a shared latent space for aligning visual and textual representations from different medical imaging fields and different languages. Inspired by contrastive learning methods, the researchers used InfoNCE (Info Noise Contrastive Estimation) loss and MSE (mean square error) loss as training objectives to maximize the similarity between positive sample pairs and minimize the similarity between negative sample pairs, thereby achieving alignment of visual representations in different fields and textual representations in different languages, laying a solid foundation for downstream zero-shot reasoning.
MultiMedLM is a module for generating multilingual reports.A multilingual text decoder was introduced, aiming to learn to generate the final medical report based on the representation extracted by MultiMedCLIP. This part is trained by reconstructing the input text, which can be Chinese text and English text, and uses the natural language generation loss - XE (cross-entropy) loss as the training target. It is worth mentioning that the introduction of reconstruction training can be regarded as unsupervised training, which only requires unlabeled plain text data for training, so there is no need to train task annotation data on downstream tasks. In addition, in order to ensure the stability of MultiMedLM training, the research team further introduced random dropout and Gaussian noise.
The experiment used the AdamW optimizer, set the learning rate to 1e-4, and the batch size to 32. The experiment was conducted on PyTorch and V100 GPU, using mixed precision training.
In terms of datasets,Pre-training was conducted on the MIMC-CXR and COVID-19-CT-CXR datasets, of which MIMC-CXR consists of 377,110 CXR images and 227,835 English radiology reports, the largest dataset released to date; COVID-19-CT-CXR includes 1k CT/CXR images and corresponding English reports. In addition, the researchers extracted half of the English corpus from the two datasets and used Google Translator to build a Chinese-English training team. The results showed that this method can improve the results of machine translation text.
During the evaluation phase, the datasets used included IU-Xray, COVID-19 CT, COV-CTR, Shenzhen Tuberculosis Dataset, COVID-CXR, NIH ChestX-ray, CheXpert, RSNA Pneumonia, and SIIM-ACR Emphysema, enabling a comprehensive evaluation of the model performance.
* IU-Xray:It includes 7,470 CXR images and 3,955 English radiology reports. The dataset is randomly divided into 80% – 10% – 10% for training, validation, and testing.
* COVID-19 CT:It contains 1,104 CT images and 368 Chinese radiology reports. The dataset is also randomly divided into 80% – 10% – 10% for training, validation, and testing.
*COV-CTR:Contains 726 COVID-19 CT images, linked to reports in Chinese and English.
* Shenzhen Tuberculosis Dataset:Contains 662 CXR images, and the training, validation and test sets are split into 7:1:2.
* COVID-CXR:Containing more than 900 CXR images, the dataset is randomly divided into 80% – 10% – 10% for training, validation, and testing.
* NIH ChestX-ray:Contains 112,120 CXR images, each image is labeled with the occurrence of 14 common radiation sickness, and the ratio of training, validation and test sets is 7:1:2.
* CheXpert:Contains more than 220,000 CXR diagnostic images. After preprocessing, 218,414 images were obtained in the training set, 5,000 images in the validation set, and 234 images in the test set.
* RSNA pneumonia:It consists of about 30k radiology images, with training, validation and test set ratios of 85% – 5% – 10%.
* SIIM-ACR Emphysema:It includes 12,047 CXR images, with the ratio of training, validation and test sets being 70% – 15% – 15% .
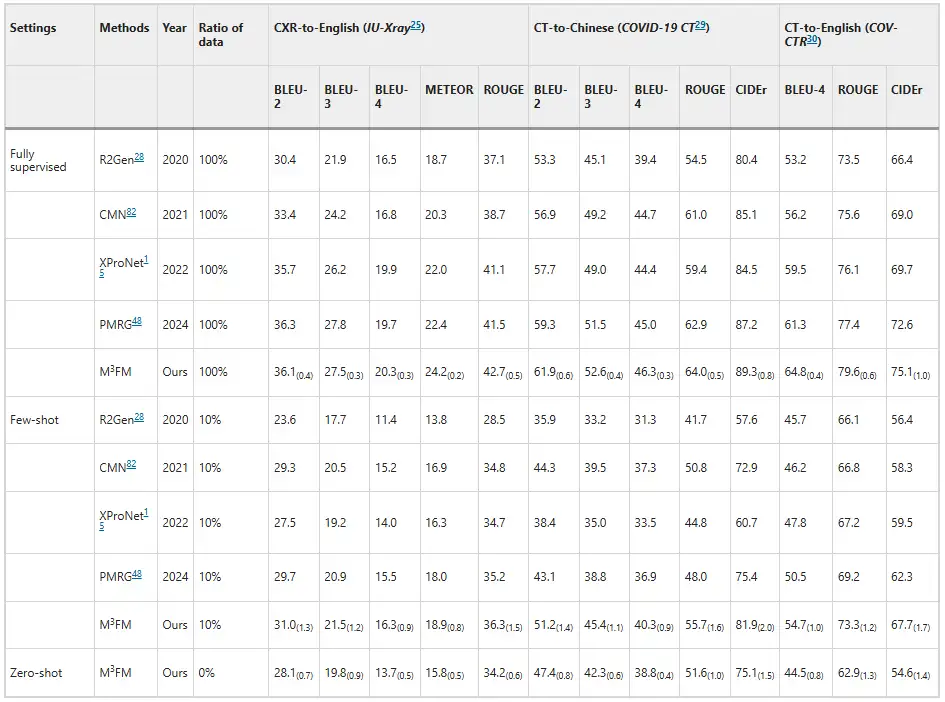
Experiments show that M³FM has superior performance, surpassing previous advanced methods.As shown in the figure below. As shown in the disease report results, in the zero-shot setting, previous methods are unable to handle the disease report task, while M³FM is able to perform multi-language, multi-domain disease reporting in a single framework. In the few-shot setting, when trained with 10% downstream labeled data, M³FM achieved the best results, even outperforming the fully supervised method R2Gen in CT-to-Chinese report generation by 1.5%'s CIDEr and 1.2%'s ROUGE-L scores.This demonstrates that M³FM can generate accurate and valid multilingual reports even when labeled data is scarce, and thus will be particularly useful for rare or emerging diseases.
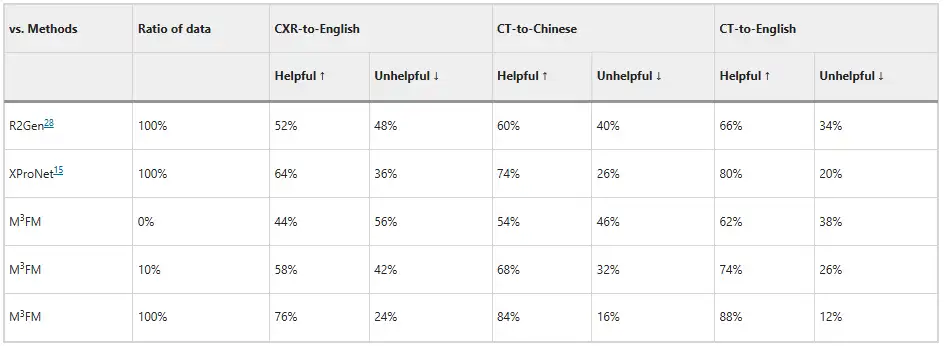
In addition, the researchers invited two clinicians to evaluate the model, and the results are shown in the figure below. Without any labeled data training, M³FM can generate ideal multilingual and multi-domain reports; when only 10% of labeled data is used for training, M³FM can be 6%, 8%, and 8% higher than the fully supervised method R2Gen in CXR-to-English, CT-to-Chinese, and CT-to-English tasks; when using complete training data, M³FM can improve R2Gen by more than 20% in three tasks, and improve XProNet by 12%, 10%, and 8% respectively.This demonstrates the potential of M³FM to free clinicians from the time-consuming and labor-intensive task of report writing.
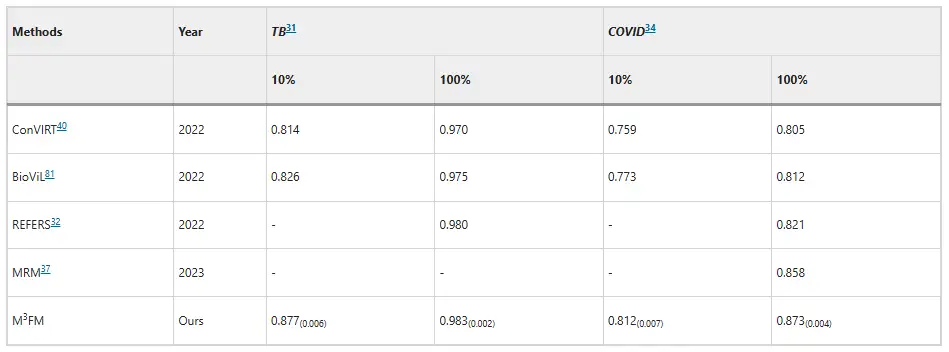
In terms of disease classification, M³FM has shown superiority in the diagnosis of infectious diseases.On the Shenzhen Tuberculosis dataset and COVID-CXR dataset, when using 10% of training data, M³FM's AUC scores are 5.1% and 3.9% higher than the existing best results, respectively. When using the training data in full, M³FM achieved the best results in two infectious diseases; in terms of non-communicable diseases, the dataset comes from NIH ChestX-ray, and M³FM achieves comparable results with the fully supervised method Model Genesis with only 1% of training labels; at 10%, M³FM outperforms the baseline methods MRM and REFERS in the diagnosis of multiple diseases, which also confirms the effectiveness and generalization ability of M³FM in disease diagnosis.
AI leads smart healthcare, and Zheng Yefeng's team takes the lead
Previously, many laboratories have focused on this issue, and the models they proposed have different focuses and advantages.
For example, for the automatic generation of reports, the School of Information Science and Technology of Dalian Maritime University published a research paper titled "DACG: Dual Attention and Context Guidance model for radiology report generation" in the professional forum Medical Image Analysis in the field of medical and biological image analysis. The paper proposed a dual attention and context guidance (DACG) model for the automatic generation of radiology reports, which can alleviate the bias of visual and text data and promote the generation of long texts.
Paper address:
https://www.sciencedirect.com/science/article/abs/pii/S1361841524003025
There are also models designed for multiple languages. For example, the team of Professor Wang Yanfeng and Professor Xie Weidi from Shanghai Jiao Tong University created a multilingual medical corpus MMedC containing 25.5 billion tokens, developed a multilingual medical question-answering evaluation standard MMedBench covering 6 languages, and built an 8B base model MMed-Llama 3, which surpassed the existing open source models in multiple benchmarks and is more suitable for medical application scenarios. The relevant research results were published in Nature Communications under the title "Towards building multilingual language model for medicine".
ClickCheckDetailed report: The benchmark test in the medical field surpasses Llama 3 and is close to GPT-4. The Shanghai Jiaotong University team released a multilingual medical model covering 6 languages
In comparison, M³FM's outstanding performance in multi-modality, multi-domain, multi-language and other aspects will undoubtedly bring new vitality to the intersection of artificial intelligence and healthcare.Of course, when talking about this research, we have to mention Dr. Zheng Yefeng, one of the authors of this article.
In fact, this paper can be said to be a fresh achievement, and it can also be seen as a sign of a new beginning for Dr. Zheng Yefeng. On July 29, 2024, IEEE Fellow, AIMBE Fellow, and medical artificial intelligence scientist Zheng Yefeng joined Westlake University full-time, was hired as a professor in the School of Engineering, and founded the Medical Artificial Intelligence Laboratory. The laboratory's research directions include medical image analysis, medical natural language understanding, and bioinformatics. This article is one of the important achievements of the laboratory's first year.
In addition to this achievement, the laboratory has also published several papers in the field of medical health, such as the research titled "Unlocking the Potential of Weakly Labeled Data: A Co-Evolutionary Learning Framework for Abnormality Detection and Report Generation", which introduces a collaborative anomaly detection and report generation (CoE-DG) framework that uses fully labeled and weakly labeled data to promote the mutual development of CXR anomaly detection and report generation tasks. The research was published in IEEE Transactions on Medical Imaging.
Of course, the lab also has research results on the currently popular large language models, such as the research titled "Mitigating Hallucinations of Large Language Models in Medical Information Extraction via Contrastive Decoding", published in EMNLP 2024. This paper provides a solution to the phenomenon that LLMs are prone to "hallucinations" in medical scenarios, and proposes an "Alternate Contrastive Decoding" (ALCD), which can significantly reduce the occurrence of errors by separating the recognition and classification capabilities of the model and dynamically adjusting the weights of the two during the prediction process. This technology performs well in multiple medical tasks.
Today, these achievements may still be in the laboratory or have the momentum to be implemented, but AI will ultimately drive the healthcare field towards intelligence, intelligence, and automation. As Dr. Zheng Yefeng said, "Medical artificial intelligence is a rapidly developing field. I estimate that in 10-15 years, artificial intelligence will have the accuracy of doctors' diagnosis and treatment and can be widely used."
References:
1.https://www.nature.com/articles/s41746-024-01339-7
2.https://mp.weixin.qq.com/s/pMNXAvzgGRpPwqVtCWjXbA
3.https://mp.weixin.qq.com/s/6hw6EJY6slAIRbGGN9XY9g