Command Palette
Search for a command to run...
Duke University Uses PepPrCLIP to Solve the "undruggable" Problem
In 2021, OpenAI released the revolutionary CLIP (Contrastive Language-Image Pre-training) model. Through unsupervised learning, CLIP can effectively understand and associate the relationship between images and text without the need for additional annotation information.
A few years later, a group of biomedical scientists were inspired by this - since CLIP matches images and language, can the same idea be used to match peptides and proteins?
By drawing on OpenAI’s breakthrough research on generating realistic images through contrastive language-image pre-training,A research team from the Department of Biomedical Engineering at Duke University constructed the PepPrCLIP (Peptide Prioritization Screening based on CLIP) pipeline, which can design short proteins (peptides) that can bind to and destroy previously undruggable disease-causing proteins.Compared to the existing platform RFDiffusion, which uses the target 3D structure to generate peptides, PepPrCLIP is faster and can create peptides that are almost always a better match to the target protein. The researchers also further verified through experiments that the "guide peptides" selected by PepPrCLIP can achieve robust and superior targeted binding and regulation in vitro as inhibitory peptides or after fusion with the E3 ubiquitin ligase domain.
The related results were published in Science Advances in January this year under the title "De novo design of peptide binders to conformationally diverse targets with contrastive language modeling".
Paper address:
https://www.science.org/doi/10.1126/sciadv.adr8638
Related dataset download address:
https://go.hyper.ai/AT5m9
The open source project "awesome-ai4s" brings together more than 200 AI4S paper interpretations and provides massive data sets and tools:
https://github.com/hyperai/awesome-ai4s
A new approach to solve the "undruggable" problem
One approach to treating disease is to develop therapeutics that can specifically target and destroy the proteins that drive the disease. Sometimes, these key proteins have well-defined structures, like carefully folded paper cranes, so conventional small-molecule therapeutics can easily bind to them.
However, pathogenic proteins with more than 80% are more like a "tangled mess", disordered and tangled together, which makes it almost impossible for standard therapies to find binding sites on their surfaces and work. The so-called "undruggable" is often used to describe proteins in traditional drug development that are difficult to become drug targets due to their structural and functional characteristics.
According to public information, difficult-to-drug targets often have the following characteristics:
* have an extended and flat functional interface, lacking a well-defined ligand binding pocket;
* Lack of specific ligands to enable the target protein to function;
* The target is a disease inhibitor, requiring a drug to activate protein activity, making drug development more challenging;
* Undruggable targets often have complex physiological functions, which increases the difficulty of drug design and development;
* Limitations of drug development strategies.
To circumvent these problems, many researchers have explored how to use peptides to bind and degrade disease-causing proteins. Since peptides are mini versions of proteins, they do not require surface pockets for binding; instead, peptides can bind to different amino acid sequences in proteins.
But even these approaches have their limitations, as existing "off-the-shelf" binders are not designed to attach to unstable or overly entangled protein structures. While scientists have been working hard to develop new binding proteins, these methods still rely on mapping the three-dimensional structural information of the target protein, which is not available for disordered targets.
The research team from the Department of Biomedical Engineering at Duke University introduced in this article took a different approach. Instead of trying to draw the structure of the disease-causing protein, they drew inspiration from large language models (LLMs) to build PepPrCLIP. Its first component, PepPr, uses a generative algorithm trained on a large library of natural protein sequences to design new "guide" proteins with specific features; the second component, CLIP, uses an algorithmic framework originally developed by OpenAI to test and screen whether these peptides can match the target protein.
Construction of a CLIP-based peptide prioritization process - PepPrCLIP
How was PepPrCLIP constructed?
Briefly, the researchers first used the ESM-2 protein language model (pLM) to perform Gaussian noise perturbations on the embeddings of real peptide binder sequences to generate candidate peptide sequences with natural characteristics; then, these candidate peptides were screened in the latent space through a CLIP-based contrastive learning architecture to train a model that co-encodes complementary peptide-protein pairs; finally, the constructed PepPrCLIP integrated a generation-discrimination framework to screen out completely novel peptide candidate sequences that are capable of binding to the target sequence.
The following figure shows the specific process of PepPrCLIP model training:
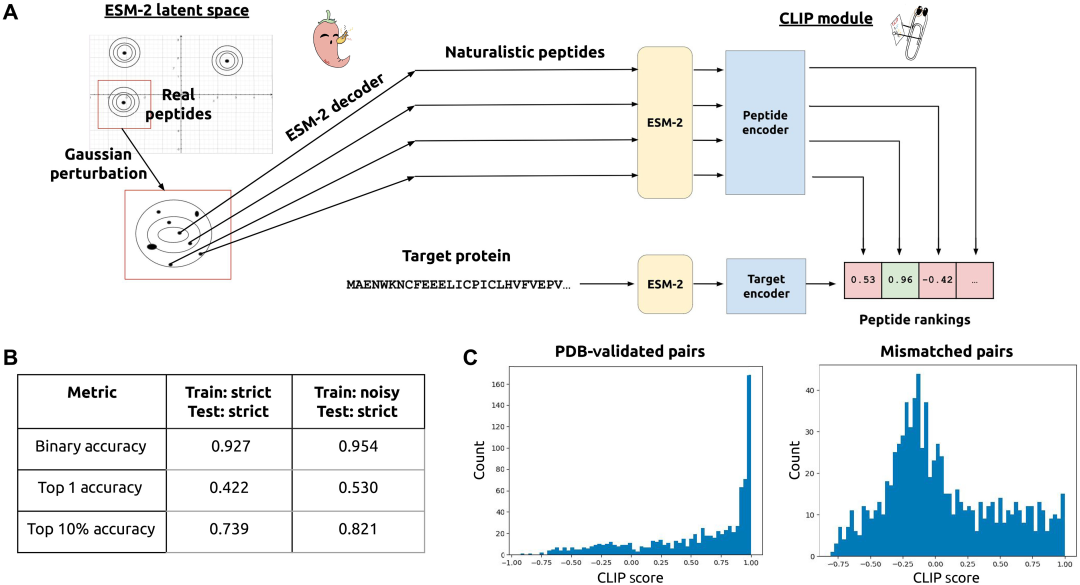
As shown in the figure above, the natural peptide embedded in ESM-2 is sampled to generate a Gaussian distribution, which is then decoded back into an amino acid sequence. The trained CLIP module jointly encodes the corresponding peptide-protein embedding, screens out thousands of peptides, and evaluates their specific binding activity to the target. Specifically:
* CLIP architecture and training
First, the input sequence is embedded through the frozen ESM-2-650M model to produce the input embedding; next, the input embedding is averaged over the sequence length to obtain an embedding vector, which is suitable for peptides and proteins; h MLP layers are applied, and the embedding vector is processed using the rectified linear unit (ReLU) activation function to obtain the output embedding. The CLIP score is obtained by performing a dot product with the vector embeddings of the peptide and protein, with a value between -1 and 1. The model is trained so that peptide-protein binding pairs have high CLIP scores.
* Generation of peptide candidate sequences
Candidate peptides are generated from all peptides in the training set, each of which is embedded using the ESM-2-650M pLM in PyTorch; for a given peptide embedding, the variance of all dimensions of the embedding is calculated; for each residue in the source peptide, noise is sampled from a standard normal distribution and multiplied by the variance to create a perturbation, which is added to the embedding of its corresponding residue. At inference time, source peptides are randomly sampled from the training set, and for each source peptide, 1,000 peptides are generated using the noise method described above. Finally, these peptides (approximately 100,000) are fed into the CLIP model and ranked based on their predicted binding to the target sequence provided by the user.
In computer simulation tests, the researchers compared the performance of PepPrCLIP with that of RFDiffusion. The researchers compared the ipTM scores of peptides generated by PepPrCLIP and those generated by RFDiffusion and found that PepPrCLIP outperformed RFDiffusion on peptides with 33% on the target, as shown in the figure below. Moreover, PepPrCLIP, which only uses sequence embedding, can greatly increase the speed of generation and prioritization, generating about 1,000 peptides per minute, and ranking 100,000 peptides per protein target in about 1 minute; in contrast, RFDiffusion takes about 2 minutes to design a single binder.This efficiency makes PepPrCLIP particularly advantageous for screening large peptide libraries, with or without structural information.
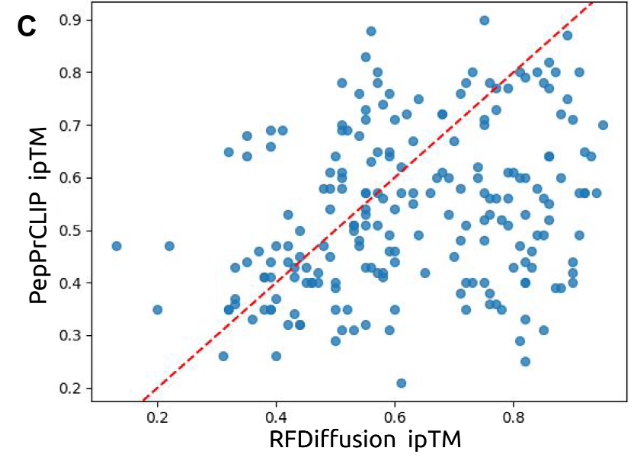
To further evaluate the effects of PepPrCLIP on ordered and disordered protein targets, the research team also collaborated with research teams at Duke University School of Medicine, Cornell University and the Sanford Burnham Prebys Medical Discovery Institute to experimentally test the platform.
In the first test, the research team showed thatPeptides generated by PepPrCLIP can effectively bind to and inhibit the activity of UltraID, a relatively simple and stable enzyme protein.
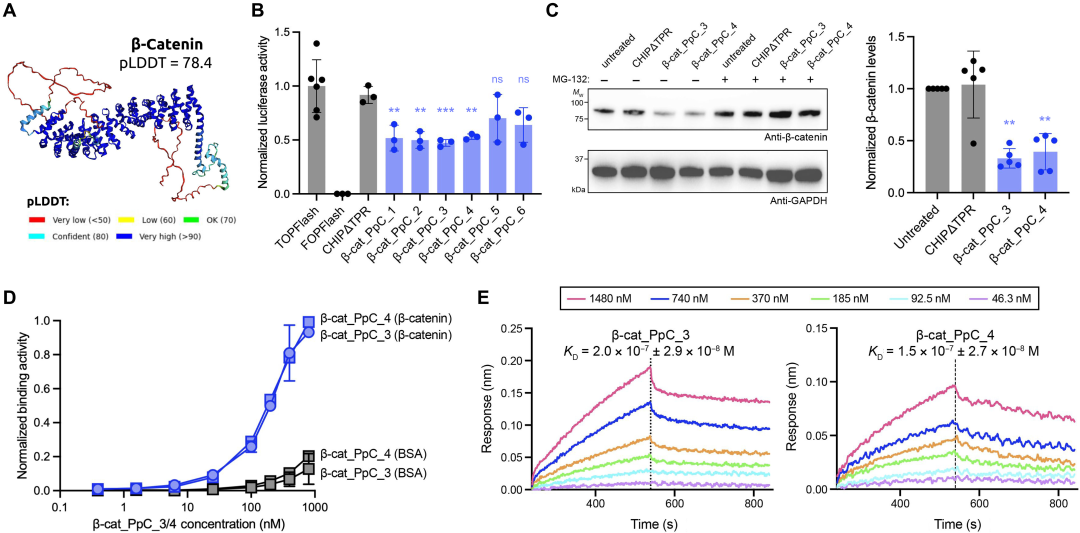
Next, they used PepPrCLIP to design peptides that could attach to β-catenin, a disordered, complex protein involved in signaling in several different types of cancer. As shown in the figure below, the team generated six peptides that CLIP showed could bind to the protein and showed that four of them could effectively bind to and degrade their targets. By disrupting the protein, they could slow cancer cell signaling.
In the most complex test, the team designed peptides that could bind to highly disordered proteins associated with synovial sarcoma, a rare, malignant cancer that accounts for 5% to 10% of all soft tissue tumors. Synovial sarcoma develops in soft tissues and primarily affects children and young adults. The disease is characterized by the presence of a unique, highly disordered oncogenic fusion protein, SS18-SSX.
The research team placed the peptides into synovial sarcoma cells and tested 10 designs. As shown in the results below, among the peptides predicted by PepPrCLIP to bind to SS18-SSX1, SS_PpC_4 significantly reduced SS18-SSX1-mCherry fluorescence; Next, the researchers also tested the effect of SS_PpC_4 overexpression on the level of endogenous SS18-SSX1 fusion protein. It is worth noting that overexpression of SS_PpC_4 peptide significantly reduced SS18-SSX1 protein levels (>40%).
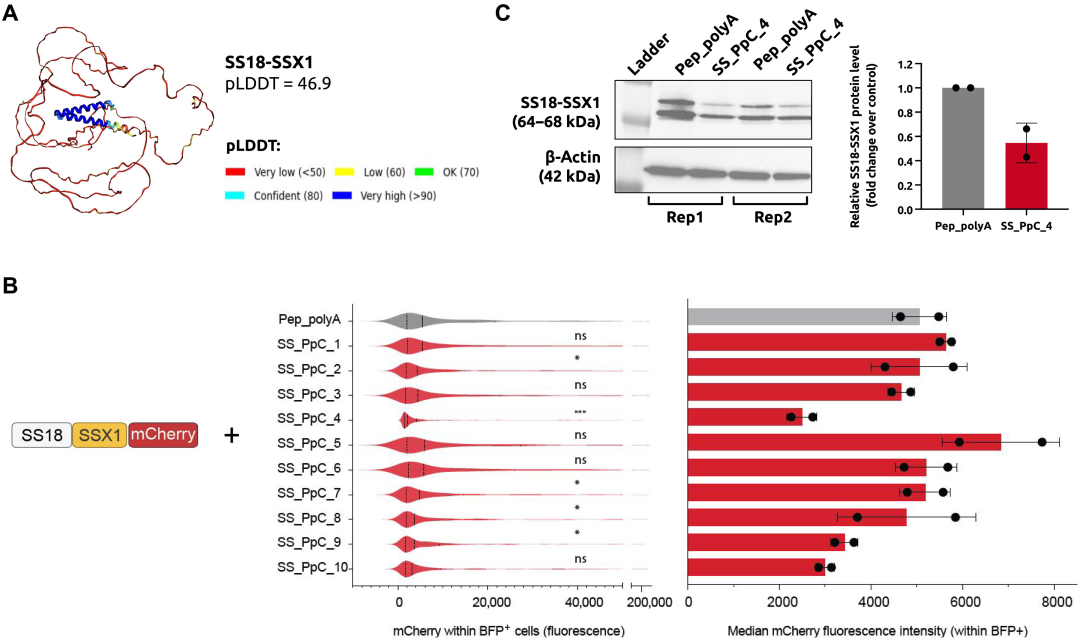
In other words,PepPrCLIP designs peptides that can both bind and degrade proteins.If they can destroy the protein, researchers have the opportunity to develop a therapy for previously undruggable cancers, which opens up many exciting clinical possibilities.
AI provides new tools to overcome "undruggable" diseases
The latest review published in a Nature journal in September 2023 comprehensively introduces the latest progress in drug discovery targeting "undruggable" proteins and its clinical application.Various molecules with similar undruggable characteristics were divided into the following categories:
① Small GTPases: such as RAS family proteins, including KRAS, HRAS and NRAS, which are considered undruggable due to the lack of targetable pockets on their surfaces;
② Phosphatase: Since each phosphatase has many similarities in structure, it has low selectivity and unavoidable side effects, which greatly hinders the progress of drug discovery.
③ Transcription factors (TFs): A variety of human diseases are associated with dysregulation of TFs involved in many biological processes, most of which cannot be targeted by conventional small molecules due to their structural heterogeneity and lack of processable binding sites.
④ Epigenetic targets: Epigenetic targets play a vital role in regulating gene expression patterns and have an impact in various biological processes and diseases;
⑤ Other proteins: Protein-protein interactions (PPIs) and their networks are of great significance in biological processes and cell cycle regulation. Some PPIs with flat interaction surfaces are more difficult to target than other PPIs, making them "undruggable" to a certain extent.
Nowadays, facing the so-called "undruggable" targets, the academic community has developed dozens of innovative methods. According to the mechanism of undruggable proteins, they have adopted fragment-based drug discovery (FBDD), computer-aided drug design (CADD), virtual screening (VS), DNA-encoded libraries (DEL) and other cutting-edge technologies to form a systematic drug design strategy. Today, the development of artificial intelligence technology and the rise of protein large language models have provided new tools for overcoming this problem.In recent years, there have been important breakthroughs in both industry and academia.
Industry,In December 2023, Absci Corporation, a leader in generative artificial intelligence antibody discovery, announced a collaboration with AstraZeneca to develop AI-designed antibodies against a tumor target. The collaboration combines Absci's Integrated Drug Creation platform with AstraZeneca's expertise in oncology to accelerate the discovery of potential new cancer treatment candidates. Absci's Integrated Drug Creation platform generates proprietary data by measuring millions of protein-protein interactions, which are used to train Absci's proprietary AI models and validate antibodies designed using new AI models in subsequent iterations. The platform accelerates drug discovery by completing data collection, AI-driven design, and laboratory validation in about 6 weeks, and is expected to expand the range of drug targets, including the development of drugs for targets previously considered undruggable.
academia,In January 2025, a study jointly led by AI pharmaceutical leader Insilico Medicine and the University of Toronto in Canada combined quantum computing models with classical computing models and generative artificial intelligence to explore a wider range of chemical possibilities through training, generation and screening of huge data sets, and discovered novel molecules targeting the "undruggable" cancer driver protein KRAS.
KRAS mutation is one of the common mutations in cancer, appearing in about a quarter of human tumors. KRAS mutation can cause uncontrolled cell proliferation and thus cause cancer. In this study, in order to generate potential new KRAS inhibitors, the researchers proposed a quantum-classical hybrid framework model that combines a quantum variational generative model (QCBM) and a long short-term memory network (LSTM), combining quantum computing with classical computing methods to design new molecules. This study was also supported by multiple research institutions such as St. Jude Children's Research Hospital. The relevant research results were published in Nature Biotechnology under the title "Quantum-computing-enhanced algorithm unveils potential KRAS inhibitors".
With the breakthroughs in related technologies, mankind has ushered in new imagination space and unlimited possibilities in defeating diseases.