Command Palette
Search for a command to run...
Huazhong University of Science and Technology Proposes a Medical Image Segmentation Model for ultra-large-scale Pathological Image Analysis to Improve the Accuracy of Sjögren's Syndrome Diagnosis
Dry mouth, dry eyes, dry skin, accompanied by unexplained muscle aches and general fatigue every day. If you experience the above symptoms, in addition to considering the dry weather in winter, you should also be alert to a common but often overlooked disease - Sjögren's Syndrome (SS).
Sjögren's syndrome is an autoimmune disease characterized by high lymphocyte infiltration of exocrine glands.About 5 million people in my country suffer from this disease. In the early stages of the disease, the exocrine glands (salivary glands, lacrimal glands, etc.) are destroyed by highly infiltrated lymphocytes, resulting in decreased function. Patients often experience dry mouth and dry eyes, and may also experience symptoms such as pain in both shoulder joints. At the same time, the disease can also affect other important organs, such as the lungs, liver, and kidneys, and even affect fertility.

Early detection and diagnosis of Sjögren's syndrome is crucial. Focal lymphocytic sialadenitis (FLS) is one of the important criteria for the diagnosis of Sjögren's syndrome. By obtaining pathological sections of the patient's minor salivary glands and performing microscopic examinations, according to existing diagnostic criteria,If more than 50 lymphocyte aggregates were found per 4 mm2 of tissue sample, it was considered to be a typical lesion.
However, a complete pathology scan image can reach 100,000*100,000 pixels, which is about 1 billion pixels. Doctors need to carefully examine the entire image and determine the number of lymphocyte aggregation foci. This is not only time-consuming, but also often relies on the experience and subjective judgment of professional doctors, increasing the risk of misdiagnosis or missed diagnosis.
To address the above challenges,Professor Tu Wei and Professor Lu Feng from Huazhong University of Science and Technology proposed the medical image segmentation model M2CF-Net using computer vision technology that is well-known in the fields of autonomous driving and facial recognition.By integrating multi-resolution and multi-scale image recognition technologies, the M2CF-Net model can not only "see" subtle differences in pathological images, but also accurately locate and count key biomarkers - lymphocyte aggregation foci, helping doctors make faster and more accurate diagnoses.
The research results were published at the 2023 IEEE International Conference on Medical Artificial Intelligence (MedAI) under the title "M2CF-Net: A Multi-Resolution and Multi-Scale Cross Fusion Network for Segmenting Pathology Lesion of the Focal Lymphocytic Sialadenitis".
Research highlights:
* Solved the problem of difficulty in identifying tiny lymphocyte clusters in ultra-large-scale tissue pathology images
* By integrating multi-resolution and multi-scale, M2CF-Net outperforms the other three mainstream medical image semantic segmentation models
* M2CF-Net performs well in processing images with blurred boundaries, small objects, and complex textures. Its segmented images have more complex shapes and are very consistent with the human-annotated ground truth

Paper address:
https://doi.ieeecomputersociety.org/10.1109/MedAI59581.2023.00063
The open source project "awesome-ai4s" brings together more than 100 AI4S paper interpretations and provides massive data sets and tools:
https://github.com/hyperai/awesome-ai4s
Dataset: Clinical data from Tongji Hospital
This study uses a set of minor salivary gland pathological section datasets from Tongji Hospital.Among them, minor salivary glands were removed from patients with primary Sjögren's syndrome.
*Minor salivary glands are distributed under the mucosa of the human oral cavity and pharynx. Their function is to secrete saliva, keep the oral cavity moist, aid digestion, and protect oral tissues from infection.
By staining the minor salivary gland pathology sections, doctors can use a microscope to observe the clear structure of the cells. Specifically, the researchers reviewed all sections to ensure their quality and confirm the presence of focal lymphocytic pharyngitis, which is characterized by more than 50 lymphocytes per 4 square millimeters around the gland. If there is a lesion, it is marked.
The final data set consists of 203 samples, including 171 positive samples (which meet the lesion characteristics) and 32 negative samples (which do not meet the lesion characteristics).The researchers divided the samples into training sets, validation sets, and test sets according to a certain ratio, which were used for model training, adjustment, and performance evaluation. In the actual process, the researchers preprocessed the data, which not only reduced the amount of calculation but also improved the generalization ability of the model.
Designing a large-scale image processing pipeline to optimize the first step of model training
The goal of this study is to extract the lesion area of focal lymphocytic sialadenitis (FLS) from the small salivary gland tissue slices with a resolution of 100,000 * 100,000. However, it is impossible to directly input gigapixel images into the neural network for training, mainly because such images are too large and the computing resources, training time, and existing frameworks are insufficient to support them.
Therefore, the researchers designed a pipeline for ultra-large-scale pathology image processing.The pipeline mainly includes three steps: Regions of Interest (ROI) extraction, stain normalization, and image patching. As shown in the following figure:
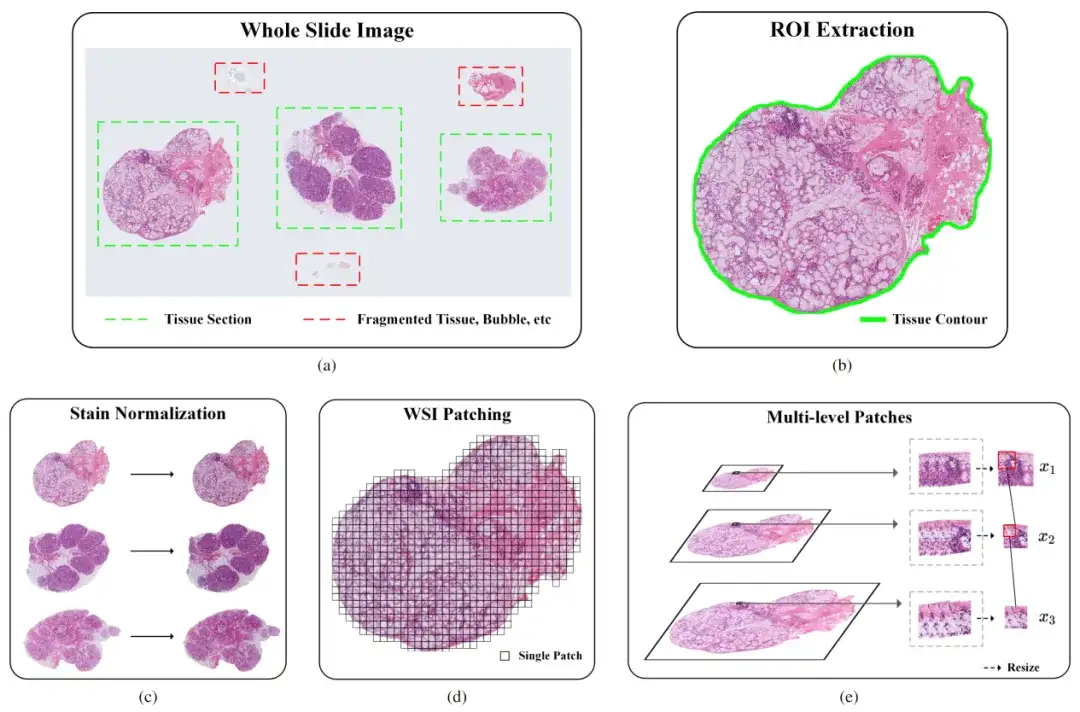
Part 1: ROI extraction
To improve the accuracy of identifying specific tissue areas in pathological images, the researchers initially used a convolutional neural network (CNN)-based classifier. However, the classifier had difficulties in dealing with complex features such as bubbles, fragmented tissue, and artifacts, resulting in its performance not meeting expectations. To address this issue, the research team took the following measures:
* Manual annotation: A portion of the samples were annotated in detail, and the classification model was retrained after enriching the dataset.
* Data augmentation: Use techniques such as rotation, scaling, and translation to increase the diversity of training data, thereby improving the accuracy of the classifier.
Part II: Staining Standardization
The main purpose of pathological image staining standardization is to ensure that images from different sources have consistent visual color and contrast. Specifically, due to factors such as stain concentration, pH value, temperature, and time, the actual staining process often causes uneven staining or inconsistent intensity, resulting in different visual effects for the same type of tissue. This difference can affect the accuracy of computer vision models.
To solve this problem, the researchers used the Vahadane algorithm. The algorithm achieves the effect of color standardization by adjusting the color characteristics of the source image to make it similar to the target image. Specifically, it calculates the color matrix transformation between the source image and the target image to achieve the color transformation of the source image.
Part III: Image Segmentation
In order to deal with the problem that the image size is still too large after ROI extraction and staining standardization, which makes it impossible to input the sample into the deep learning model for training, the researchers adopted a patch-based training method to divide the image into small patches with overlapping areas, which not only improves the model training efficiency but also retains the original information.
In order to analyze the detailed features of small lymphocytes near larger ducts, it is necessary to capture tissue-level features in a larger field of view. However, in order to ensure the accuracy of the segmentation results, it is necessary to capture cell-level features in a smaller field of view. How to balance the relationship between the two is particularly important.
To this end, the researchers considered a multi-resolution image segmentation method, which mainly involves downsampling the original image several times and extracting image blocks of the same size from these downsampled images. These patches cut from images with different sampling magnifications have different fields of view, which can capture both tissue-level features and cell-level features.
Multi-resolution and multi-scale fusion model, efficient performance improvement
The model M2CF-Net selected by the researchers includes a multi-branch encoder and a fusion-based cascade decoder.The encoder downsamples features of patches of different resolutions at different scales, while the decoder uses a cascaded fusion block to fuse feature maps generated by multi-branch encoders.
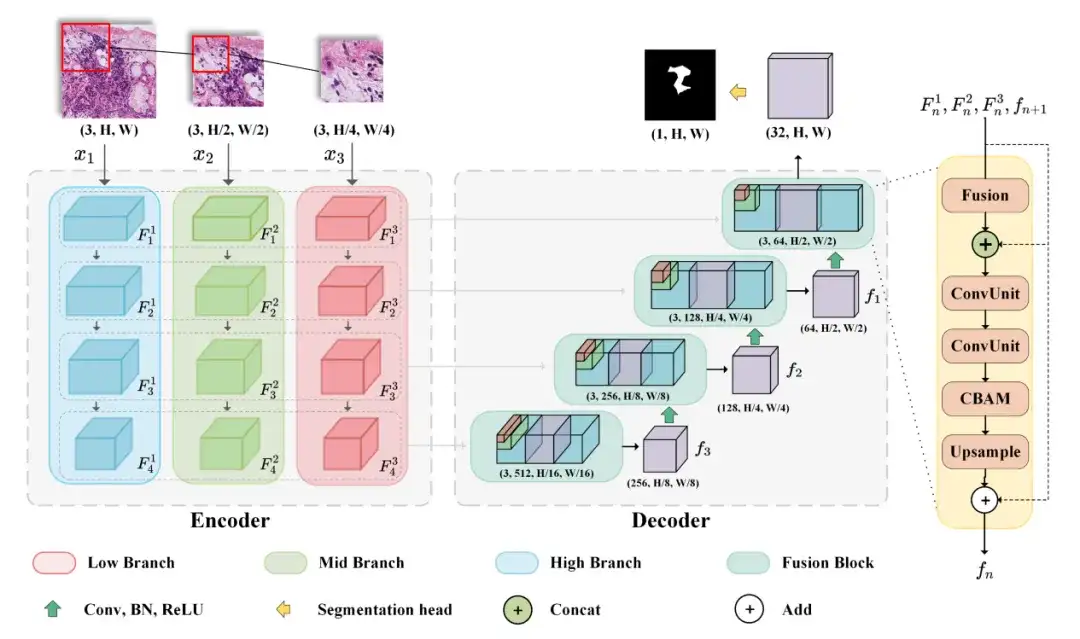
Specifically, in order to obtain features at both the tissue level and the cell level, the researchers designed a multi-branch network, which is a typical Encoder-Decoder architecture model that can accept images of different resolutions as input. The Encoder includes three input branches, which accept images of different resolutions and generate feature map combinations of different field of view during the encoding process. The Decoder can fuse the feature map combinations generated by the Encoder using cascaded Fusion Blocks to output the final prediction map.
In this process, the researchers also used spatial attention and channel attention mechanisms to enhance the representation ability of input features. Finally, BCEDice Loss was used as the loss function of the model, which weighed the binary cross entropy loss and Dice loss and can effectively guide the optimization direction of the model.
Experimental conclusion: M2CF-Net outperforms the other three mainstream medical image semantic segmentation models
The researchers compared their proposed model (M2CF-Net) with four other popular medical image semantic segmentation models - UNet, MSNet, HookNet and TransUNet. The results showed that the M2CF-Net model has more advantages in utilizing multi-resolution and multi-scale features.
* UNet: uses an encoder-decoder structure to capture multi-scale features for accurate segmentation
* MSNet: Introduces a multi-scale subtraction network to enhance feature extraction and improve segmentation accuracy
* HookNet: Hook is added to capture and utilize multi-resolution features, enhance the U-Net structure, and effectively handle the segmentation of images of various sizes in medical images
* TransUNet: Based on Transformer, it improves segmentation accuracy by introducing a self-attention mechanism
As shown in the figure below, the researchers found that M2CF-Net achieved the highest Dice of 69.40%, and its number of parameters was only half of that of TransUNet, which ranked third. It outperformed UNet and MSNet, which had fewer parameters, by 38.9% and 22.5%, respectively.It can effectively capture and fuse features of different scales in images.
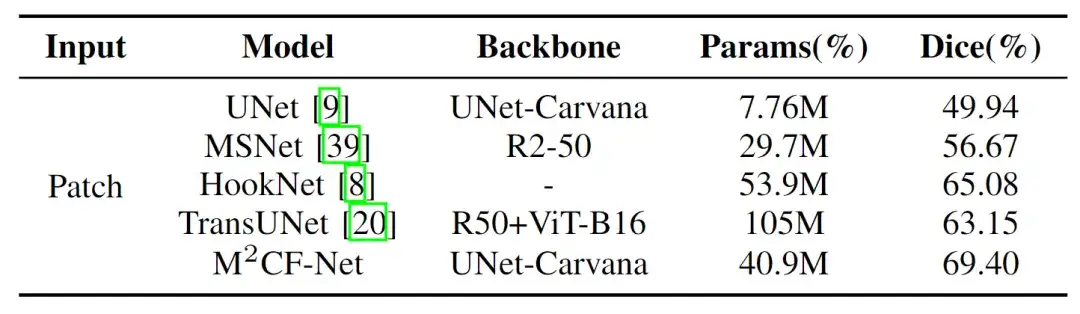
Specifically, the number of parameters (Params) of M2CF-Net is less than TransUNet and HookNet, but more than UNet and MSNet. This is because TransUNet is based on the Transformer architecture and has more parameters than CNN, and the single-branch decoder makes the number of parameters of M2CF-Net less than HookNet. However, compared with the single-branch input network, the multi-branch encoder structure in M2CF-Net leads to a higher number of parameters.
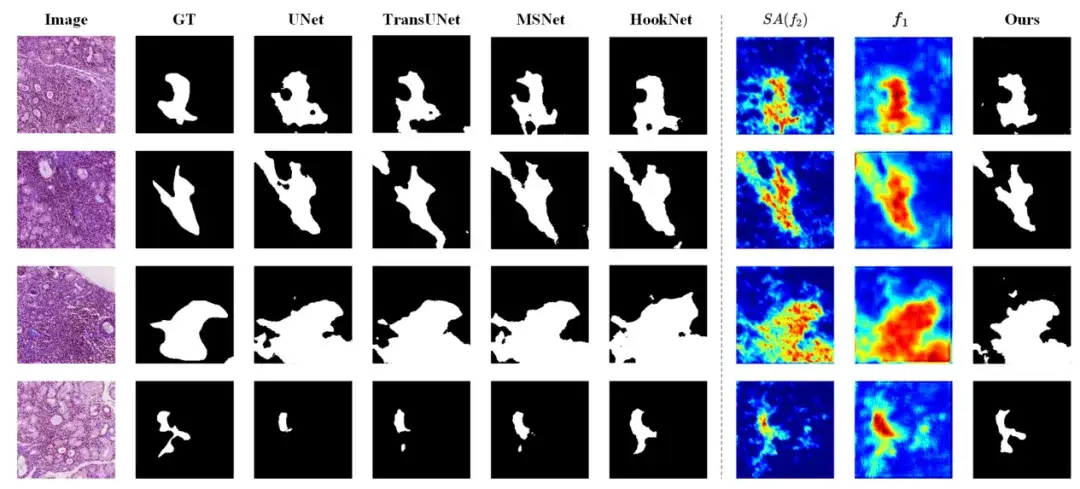
In addition, after in-depth analysis,The study found that M2CF-Net performs well in processing images with blurred boundaries, small objects, and complex textures. As shown in the figure below, the segmentation results of M2CF-Net have more complex shapes, which is very consistent with the actual human annotations.
Computer vision technology revolutionizes medical image segmentation
Medical image analysis is crucial for disease diagnosis. Computer technology can be used to accurately segment medical images and effectively identify lesions, human organs, and infected areas, thereby improving diagnostic efficiency. In recent years, thanks to the advancement of advanced technologies such as deep learning, medical image segmentation technology is rapidly shifting from manual operations to automated processing. Specially trained AI systems have now become an indispensable auxiliary tool for medical professionals.
Professor Tu Wei, Deputy Director of the Department of Rheumatology and Immunology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology,With more than 20 years of experience in the diagnosis and treatment of rheumatic and immune diseases, he has extensive experience in the diagnosis of Sjögren's syndrome. In this study, Professor Tu Wei deeply analyzed the pathological diagnosis process of Sjögren's syndrome, pointed out the key points that are easy to confuse, and demonstrated the diagnostic results in different situations through actual cases. After mastering the pathological diagnosis method of Sjögren's syndrome,Professor Lu Feng's team proposed using image segmentation technology in computer vision to address diagnostic challenges.The two parties have used advanced AI technology to open up a new path for the diagnosis of Sjögren's syndrome.
Professor Tu Wei's personal homepage:
https://www.tjh.com.cn/MedicalService/outpatient_doctor.html?codenum=101110
Professor Lu Feng's personal homepage:
http://faculty.hust.edu.cn/lufeng2/zh_CN/index.htm
In addition to the researchers mentioned above, there are many scientists dedicated to cutting-edge research in the intersection of medical imaging and AI.
For example, a team from the Massachusetts Institute of Technology's Computer Science and Artificial Intelligence Laboratory (MIT CSAIL), in collaboration with researchers from Massachusetts General Hospital and Harvard Medical School, proposed a general model, ScribblePrompt, for interactive biomedical image segmentation.This neural network-based segmentation tool not only supports annotators using different annotation methods such as scribbles, clicks, and bounding boxes to perform flexible biomedical image segmentation tasks, but also performs well for untrained labels and image types.
I believe that as more advanced technologies are developed and applied in clinical practice, multiple medical branches such as oncology and neurology will benefit, and the field of medical image analysis will usher in a brighter development prospect.