Command Palette
Search for a command to run...
Comparing 11 Algorithms Horizontally, the University of Toronto Launched a Machine Learning Model to Accelerate the Development of New long-acting Injectable Drugs
According to the "Report on the Nutrition and Chronic Disease Status of Chinese Residents (2020)", in 2019, deaths due to chronic diseases accounted for 88.51% of the total deaths in my country.Chronic diseases have become a major "killer" threatening human health.Take schizophrenia, which is called "the worst disease of mankind" by scholars, for example. If patients want to fully recover, they need to undergo long-term maintenance treatment. However, during this period, patients may stop taking medication for various reasons, resulting in relapse.
In order to solve the problem of poor medication compliance among chronic patients, long-acting injections have been introduced. This drug is a drug that is dissolved in a certain preparation with a sufficient dose, injected into the body to form a small drug "storage warehouse", and then slowly releases the drug in the body to play a stable therapeutic role. Compared with traditional drugs,Long-acting injections have the advantages of long dosing intervals, rapid action, and stable drug dosage.
On the other hand, the development of this new type of drug is also quite challenging. For example, in order to achieve the optimal release of the drug in the body within a specified time frame, a large number of extensive experiments are required on a variety of candidate preparations. This process is cumbersome and time-consuming, and has become a bottleneck for the further development of long-acting injections.
Recently, researchers from the University of Toronto developed a machine learning model. Relevant experimental results show that the model can accurately predict the release rate of long-acting injectable drugs, effectively accelerating the development of long-acting injectable drugs.The research has been published in the journal Nature Communications.The title is "Machine learning models to accelerate the design of polymeric long-acting injectables".

The results have been published in Nature Communications.
Paper address:
https://www.nature.com/articles/s41467-022-35343-w#Abs1
Experiment Overview
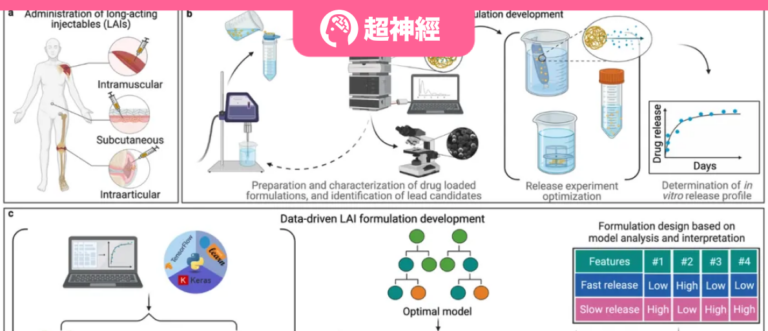
Long-acting injectable preparations come in a variety of types, generally lipids and synthetic polymers.The figure below shows a comparison between traditional and data-driven approaches to the development of long-acting injectable formulations.

Figure 1: Schematic diagram of traditional and data-driven R&D approaches for long-acting injectable formulations
a Figure: Routes of administration of long-acting injectable formulations approved by the US Food and Drug Administration.
Figure b: Typical trial-and-error cycle in the development of traditional long-acting injectable formulations.
Figure c: Overview of the workflow of this study, which uses trained machine learning models to accelerate the development process of long-acting injectable formulations.
This experimental dataset is constructed from previously published research results.At the same time, data from external sources searched from the Web of Science engine were also added. Specifically, the data set includes the release amounts (the number of drug molecules released in a given time) of 181 drugs and 43 drug-polymer combinations. At the same time, the researchers divided the constructed data set into two subsets:They are used for model training and testing respectively.
Long-Acting Injectables Dataset
Publishing Agency:University of Toronto
Quantity included:Release of 181 drugs and 43 drug-polymer combinations
Estimated size:394.1 KB
Release time:2022
Download address:hyper.ai/datasets/23625
Experimental procedures
In this study, the researchers trained a total of 11 machine learning algorithms.Including multiple linear regression (MLR), least absolute shrinkage and selection operator (Lasso), partial least squares regression (PLS), decision tree (DT), random forest (RF), light gradient boosting machine (LGBM), extreme gradient boosting (XGB), natural gradient boosting (NGB), support vector regression (SVR), k nearest neighbor algorithm (k-NN) and neural network (NN).
Model selection
To evaluate the predictive performance of these machine learning models, the researchers used a nested cross-validation approach that consisted of an inner (training and validation) and outer (testing) loop. The researchers first grouped the dataset by drug-polymer combination,Then, 10 nested cross-validation experiments were performed on each machine learning model.
Finally, the prediction performance of each machine learning model in the inner and outer nested cross-validation loops is summarized in Table 1 and Figure 2. Table 1 shows the mean absolute error (MAE) values and mean standard error (σM, shown in brackets) obtained after predicting drug release using different machine learning algorithms in nested cross-validation (n=10). As can be seen from the table,Tree-based machine models are generally more accurate than linear, instance-based, and deep learning models (MAE < 0.16).

Table 1: Prediction performance of each machine learning model in nested cross validation
Figure 2 shows the absolute error (AE) values of drug release prediction obtained in the nested cross-validation (n=10). Combining the information in Table 1 and Figure 1, the LGBM-based model has the smallest MAE and AE values in the inner and outer loops among the 11 models.The researchers believe that the LGBM-based model is the one with the best predictive performance.

Figure 2: Overall prediction performance of each algorithm model
The black circles and black dashed lines in the boxes in the figure represent the MAE value and AE value of each model, respectively.
Model Optimization
In order to further improve the generalization ability of machine learning models,The researchers also optimized and improved the LGBM model with 17 features through cluster analysis.
Here they used the farthest neighbor clustering algorithm, as shown in the figure below, to arrange the input features into a hierarchical structure. The researchers found that there was redundancy in 17 features. After improvement,finalIt was determined that the LGBM model with 15 features performed best.

Figure 3: Spearman correlation coefficient heat map of the initial 17 input features
Dark blue indicates an absolute Spearman correlation coefficient of 1, and pink indicates an absolute Spearman correlation coefficient of 0. Next to the heat map is a tree diagram showing the hierarchy of feature clusters determined by agglomerative hierarchical cluster analysis.
Experimental Results
After obtaining the above optimal model, the researchers conducted two tests. One was to use the model to predict the drug release curve of a certain long-acting injection drug, and the other was to use the model to predict the drug release curve of the drug-polymer in the test set. The results were compared with the experimental drug release curves, and the results are shown in the figure below.
Figure 4 shows the comparison of predicted and experimental drug release profiles for a selected long-acting injection, and Figure 5 shows the comparison of drug release profiles for drug-polymer and experimental drug release profiles. It can be seen that in both cases,The predicted values and experimental values are basically consistent.Therefore, the researchers believe that the model based on the LGBM algorithm can accurately predict the drug release rate of long-acting injections.

Figure 4: Comparison of predicted and experimental drug release curves for long-acting injections in the dataset

Figure 5: Comparison of drug-polymer predicted and corresponding experimental drug release profiles
Acceleration Alliance: Helping the implementation of new scientific research paradigms
It is worth noting thatThe authors of this research, Christine Allen and Alán Aspuru-Guzik, are both from the Acceleration Consortium (AC).Born in 2021, the Accelerator Alliance is a new global collaboration between academia, industry and government, headquartered at the University of Toronto, Canada, with the vision of using AI and robotics to accelerate the discovery and design of new materials and molecules.
"Our goal is to accelerate science,"“To achieve this goal, we realized we could extend the thinking behind autonomous driving to automated labs, using AI and automation to conduct experiments in smarter ways,” said Alán Aspuru-Guzik, director of the Accelerator Alliance.

Figure 6: Accelerator Alliance, a scientist takes out pre-dispensed reagents from an automated solid dispensing robot
It is worth noting that just last month, the Accelerator Alliance received a grant of $200 million from the Canada First Research Excellence Fund (CFREF), which will be used to support the alliance's work in the field of "self-driving labs". In this regard, Meric Gertler, president of the University of Toronto, said,"These significant investments in AI-driven research and innovation have the potential to improve the lives of people in Canada and around the world.".
The code address of this study:
https://github.com/aspuru-guzik-group/long-acting-injectables