Command Palette
Search for a command to run...
Selected for ACL2024 Main Conference | InstructProtein: Aligning Protein Language With Human Language Using Knowledge Instructions

As the basis for cell survival, protein exists in all organisms including the human body. It is the scaffold and main substance that constitutes tissues and organs, and plays a core role in chemical reactions that are essential to life.
Faced with the complexity and variability of protein structure, traditional experimental methods are time-consuming and laborious in analyzing protein structure, so protein language models (PLMs) came into being. These professional models use amino acid sequences as input and can predict protein functions and even design new proteins. However,Although PLMs are excellent at understanding amino acid sequences, they are unable to understand human language.
Similarly, when large language models (LLMs) such as ChatGPT and Claude-2, which are good at processing natural language, are asked to describe the function of protein sequences or generate proteins with specific properties, they are also unable to do so.There are two major deficiencies in the current protein-text pair datasets: one is the lack of clear instruction signals; the other is the imbalance of data annotation. In short, there is an unresolved gap in the current research on LLMs, which is the inability to quickly convert between human language and protein language.
To solve this kind of problem,The team led by Huajun Chen and Qiang Zhang from Zhejiang University proposed the InstructProtein model, which uses knowledge instructions to align protein language with human language.We explored the bidirectional generation capabilities between protein language and human language, effectively bridging the gap between the two languages and demonstrating the ability to integrate biological sequences into large language models.
The research is titled "InstructProtein: Aligning Human and Protein Language via Knowledge Instruction".Accepted by ACL 2024 main conference.
Research highlights:
* InstructProtein is a study to align human language and protein language through knowledge instructions
* Explored the bidirectional generation capabilities between protein language and human language, effectively bridging the gap between the two languages
* Experiments on a large number of bidirectional protein-text generation tasks show that InstructProtein outperforms existing state-of-the-art LLMs

Paper address:
https://arxiv.org/abs/2310.03269
The open source project "awesome-ai4s" brings together more than 100 AI4S paper interpretations and provides massive data sets and tools:
https://github.com/hyperai/awesome-ai4s
Dataset: Comprehensive scientific dataset
The corpus for the model pre-training phase contains protein sequences from UniRef100 and sentences from PubMed abstracts.Based on this data, the researchers generated an instruction dataset containing 2.8 million data points.
During the fine-tuning stage of the model, the protein knowledge graph was constructed using the annotations provided by UniProt/Swiss-Prot, which included protein superfamilies, families, domains, conserved sites, active sites, binding sites, locations, functions, and biological processes involved; the data for knowledge causal modeling came from the InterPro and Gene Ontology databases.
In the model evaluation stage, the researchers selected the Gene Ontology (GO) dataset to evaluate the model's ability in protein function annotation, and then selected the Hu et al. dataset to evaluate the model's ability in metal ion binding (MIB) prediction.
Model architecture: Fine-tuning the pre-trained model by building a protein knowledge instruction dataset
To give LLM the ability to understand protein language, InstructProtein adopts a two-step training approach: first pre-training on protein and natural language corpora, and then fine-tuning with an established protein knowledge instruction dataset.
Pre-training phase
In the multilingual pre-training stage, this study uses a large biological text database to enhance the model's language understanding and knowledge background in the biological field. Multilingual refers to the ability to process natural language (such as English abstracts) and biological sequence language (such as protein sequences).
Model fine-tuning phase
During the model fine-tuning phase,This study proposes a dataset construction method called "knowledge instruction".The knowledge graph (KGs) and the large language model work together to build a balanced and diverse instruction dataset. This method does not rely on the large language model's ability to understand protein language, thus avoiding false information introduced by model bias or hallucination. The specific construction process is divided into 3 main stages, as shown in the figure below:
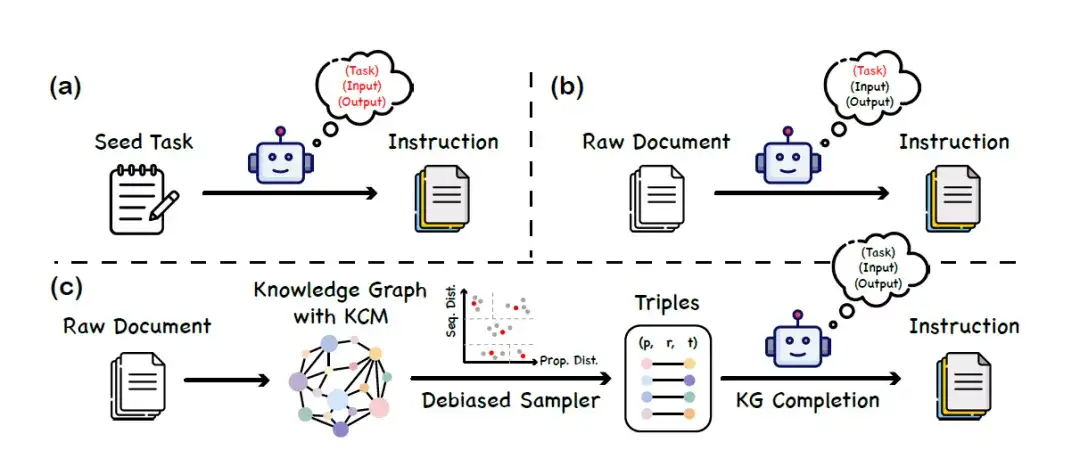
a. Given a set of seed tasks, prompt LLM to generate new instruction data
b. Use LLM to generate instruction data corresponding to the content of the raw document
c. Instruction generation framework based on knowledge graph (KG)
* Construction of knowledge graph:The researchers used UniProtKB as a data source to build a protein knowledge graph. Drawing on the concept of chain thinking, the researchers realized that there are also logical chains in protein annotations. For example, the biological processes that proteins can participate in are closely related to their molecular functions and subcellular localization, and the molecular functions themselves are affected by the protein domains.
To represent the causal chain of this protein knowledge,The researchers introduced a new concept called Knowledge Causal Modeling (KCM).Specifically, the knowledge causal model consists of multiple interconnected triplets organized in a directed acyclic graph, where the direction of the edge represents the causal relationship. The graph organizes the triplets from the micro level (covering the characteristics of the protein sequence, such as structure) to the macro level (covering biological functions). The figure below shows the process of generating factual, logical, and diverse instructions using a large language model combined with a knowledge graph to complete the task given a triple containing a KCM.
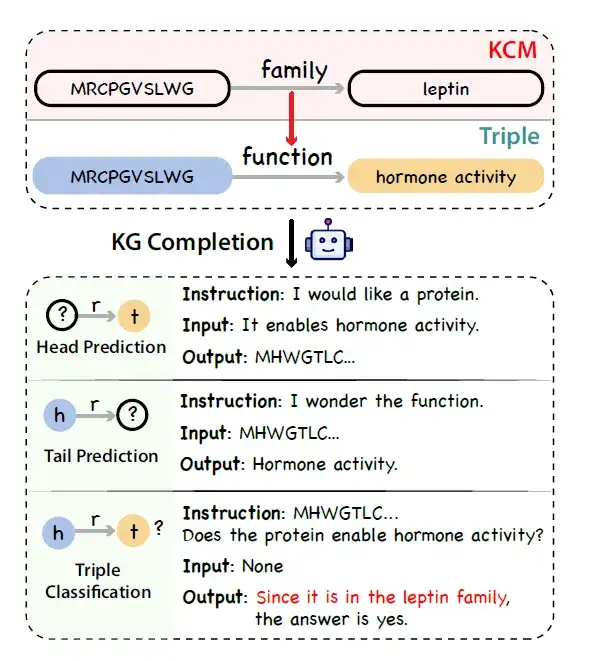
* Knowledge graph triple sampling:Considering the annotation imbalance problem in knowledge graphs, the researchers proposed a debiased sampling strategy to replace uniform sampling as an alternative to unified sampling. Specifically, they first grouped proteins according to their sequence and attribute similarities, and then uniformly extracted triplets in each group.
* Generation of instruction data:The researchers simulated the knowledge graph completion task and used a general LLM (such as ChatGPT) to convert knowledge graph triples with KCM into instruction data.
This approach allows for the efficient creation of a rich and balanced dataset of instructions for protein function and location without relying on pre-defined models to understand protein language.Provide more reliable data support for subsequent protein function research and application.
Through a combination of pre-training and fine-tuning, the resulting model, called InstructProtein, is able to better perform various prediction and annotation tasks involving protein sequences.For example, accurately predicting the function of a protein or localizing it to a specific subcellular location - which has important implications for protein engineering, drug discovery, and broader biomedical research.
Research results: InstructProtein outperforms existing state-of-the-art LLMs
The study comprehensively evaluated InstructProtein's capabilities in protein sequence understanding and design:
Protein sequence understanding
The researchers evaluated the performance of the InstructProtein model on the following three classification tasks:Protein location prediction, protein function prediction, protein metal ion binding ability prediction. These tasks are designed to be similar to reading comprehension problems in natural language, where each piece of data contains a protein sequence and a question, and the model needs to answer yes/no questions. All evaluations are performed in a zero-shot setting.
The evaluation results are shown in the following table:InstructProtein achieves new state-of-the-art performance on all tasks compared to all baseline models.
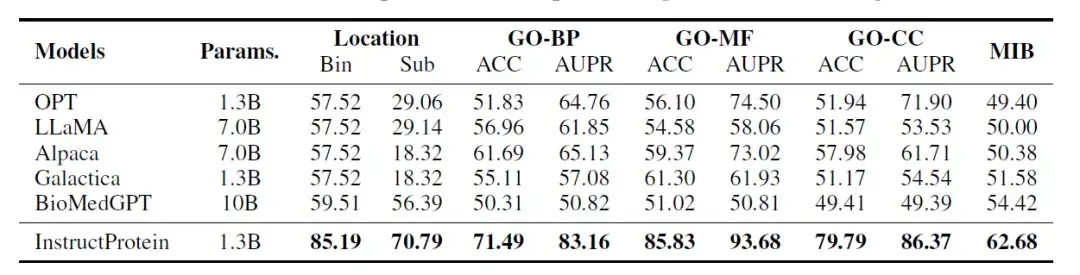
In addition, two key findings are worth noting. First, InstructProtein significantly outperforms LLMs derived from natural language training corpora (i.e., OPT, LLaMA, Alpaca). This suggests thatTraining with a corpus containing both proteins and natural language is beneficial for LLMs and improves their capabilities in protein language understanding.
Second, although both Galactica and BioMedGPT use UniProtKB as the corpus for natural language and protein alignment, InstructProtein consistently outperforms them.The high-quality instruction data in this study can improve the performance in zero-shot settings.
In addition, in the protein subcellular localization (bin) task, LLMs (OPT, LLaMA, Alpaca, and Galactica) were severely biased, causing all proteins to be classified into the same group, resulting in an accuracy of 57.52%.
Protein sequence design
In terms of protein design, the researchers designed an "instruction protein pairing" task: given a protein and its description, the model needs to select the most appropriate one from its corresponding description and 9 non-corresponding descriptions.
As shown in the following table:InstructProtein significantly outperforms all baseline models in the instruction-protein pairing task.
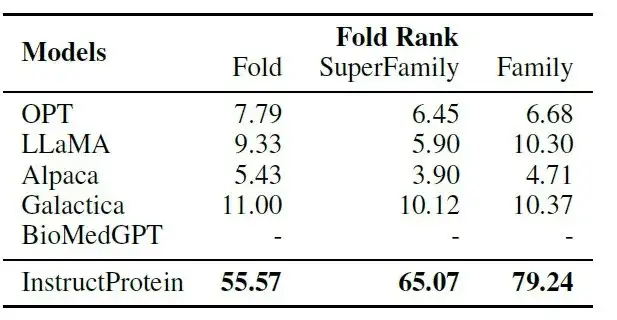
Among them, BioMedGPT focuses on converting proteins to text and lacks protein design capabilities; Galactica has limited performance in the zero-shot setting of aligning instructions with proteins because it is trained on a narrative protein corpus.These results confirm the superiority of the InstructProtein model's instruction-following capabilities in protein generation.
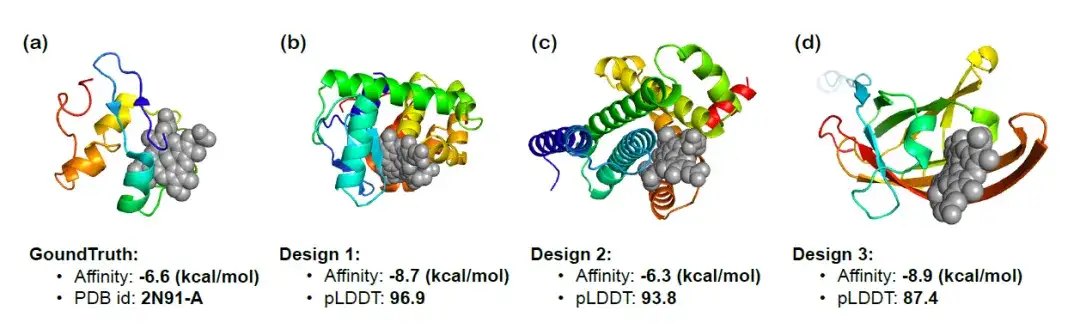
To further verify InstructProtein's ability to design proteins according to function-related instructions, the researchers used InstructProtein to design heme-binding proteins that can bind to specific compounds and visualized the 3D structures of the three generated proteins. The figure below shows the docking results, binding affinity prediction (the lower the better), and pLDDT score (the higher the absolute value, the better). It can be observed that the generated proteins show significant binding affinity,The effectiveness of InstructProtein in the design of heme-binding proteins was confirmed.
The road to exploring protein models has just begun
In recent years, large language models have revolutionized the field of natural language processing. These models are widely used in many aspects of daily life, such as language translation, information acquisition, and code generation. However, although these language models perform well in processing natural and code languages, they are unable to cope with biological sequences (such as protein sequences) -In this context, the emergence of protein large language model is timely.
Protein language models are specifically trained on protein-related data, including amino acid sequences, protein folding patterns, and other biological data related to proteins. Therefore, they have the ability to accurately predict protein structure, function, and interaction. Protein language models represent the cutting-edge application of AI technology in biology. By learning the patterns and structures of protein sequences, they can predict the function and morphology of proteins, which is of great significance for new drug development, disease treatment, and basic biological research.
In April 2023, a study published in Science showed that researchers from the meta AI team used a large language model that can emerge evolutionary information to develop a sequence-to-structure predictor ESMFold. The prediction accuracy for single-sequence proteins exceeded that of AlphaFold2, and the prediction accuracy for proteins with homologous sequences was close to that of AlphaFold2, and the speed was increased by an order of magnitude. The model predicted more than 600 million metagenome proteins, demonstrating the breadth and diversity of natural proteins.
In July 2023, Baidu Biosciences and Tsinghua University jointly proposed a model called xTrimo Protein General Language Model (xTrimoPGLM), which has 100 billion parameters. In terms of understanding tasks, xTrimoPGLM significantly outperforms other advanced baseline models in a variety of protein understanding tasks; in terms of generation tasks, xTrimoPGLM can generate new protein sequences similar to natural protein structures.

Paper link:
https://www.biorxiv.org/content/10.1101/2023.07.05.547496v3
July 2024Zhou Hao, an associate researcher at the Institute of Intelligent Industries of Tsinghua University, worked with Peking University, Nanjing University and Shuimu Molecular Team to propose a multi-scale protein language model ESM-AA (ESM All Atom).By designing training mechanisms such as residue expansion and multi-scale position encoding, the ability to process atomic-scale information has been expanded. The performance of ESM-AA in tasks such as target-ligand binding has been significantly improved, surpassing the current SOTA protein language models, such as ESM-2, and also surpassing the current SOTA molecular representation learning model Uni-Mol. Related research has been published at the top machine learning conference ICML under the title "ESM All-Atom: Multi-scale Protein Language Model for Unified Molecular Modeling".

Paper address:
https://icml.cc/virtual/2024/poster/35119
It is worth emphasizing that although significant progress has been made in the study of protein large language models, we are still in the early stages of fully grasping the complexity of protein sequence space. For example, the InstructProtein model mentioned above has challenges in handling numerical tasks, which is particularly important in the field of protein modeling that requires quantitative analysis, including the establishment of 3D structure, stability assessment, and functional evaluation. In the future,Related research will be extended to include a wider range of instructions including quantitative descriptions,Enhance the model's ability to provide quantitative output, thereby advancing the integration of protein language and human language and expanding its practicality in different application scenarios.
References:
1.https://arxiv.org/abs/2310.03269
2.https://mp.weixin.qq.com/s/UPsf9y9dcq_brLDYhIvz-w
3.https://hic.zju.edu.cn/ibct/2024/0228/c58187a2881806/page.htm