Command Palette
Search for a command to run...
Live Preview: Tsinghua University Postdoctoral Fellow Li Yuzhe Explains Cell/Nature sub-journal Papers and Explores AI Applications in Genomics

The second episode of the "Meet AI4S" live series will be broadcast at 19:00 on August 21! HyperAI is honored to invite Li Yuzhe, a postdoctoral fellow in Zhang Qiangfeng's laboratory at Tsinghua University, to share AI methods in spatial transcriptomics and single-cell omics research.
The research results of Dr. Li Yuzhe as the first author, "Tissue module discovery in single-cell resolution spatial transcriptomics data via cell-cell interaction-aware cell embedding", have been published in Cell Systems.He will also further introduce the innovative method proposed in this study: the artificial intelligence algorithm SPACE based on the graph autoencoder deep learning framework.
also,Dr. Li Yuzhe participated in the research of artificial intelligence algorithms based on the variational autoencoder deep learning framework SCALEX,The paper was published in Nature Communications with the title "Online single-cell data integration through projecting heterogeneous datasets into a common cell-embedding space". He will also share the research ideas of this achievement with everyone in the live broadcast.
Event Details

Share the topic
Exploring AI applications in genomics: Taking the spatial transcriptome data representation algorithm SPACE as an example
Introduction
With the rapid development of life science research and artificial intelligence (AI) technology, AI is playing an increasingly important role in biomedical research, giving rise to a new cross-disciplinary research paradigm of "AI for Science".
This live broadcast will share genomics research, mainly artificial intelligence methods in spatial transcriptomics and single-cell omics research.
Audience benefits
1. Gain a preliminary understanding of the development process and cutting-edge progress of artificial intelligence methods in genomics research.
2. Understand SCALEX, a single-cell omics data integration algorithm based on generative models.
3. Understand the spatial transcriptomics data representation algorithm SPACE based on graph neural network and the concept of cell community.
Paper Review
HyperAI has previously interpreted and shared a research paper titled "Tissue module discovery in single-cell resolution spatial transcriptomics data via cell-cell interaction-aware cell embedding" with Dr. Li Yuzhe as the first author.
Research highlights
* Developed SPACE, an artificial intelligence analysis tool for spatial transcriptome data, which can identify spatial cell types and discover tissue modules from spatial transcriptome data with single-cell resolution.
* SPACE significantly outperforms other tools in cell type identification and tissue module discovery, especially in complex tissues containing multiple cell types.
* SPACE defines and discovers cell communities, i.e., tissue modules composed of spatially homogeneous cell types with recognizable boundaries.
* Cell communities are defined by similar interaction networks among the cells they comprise, which can be used to refine ligand-receptor based inferences about cell communication.
* SPACE can be used for large-scale spatial transcriptome studies to understand how interactions between spatially neighboring cells affect the biological functions of cell types and tissue modules.
Dataset acquisition
In order to verify the capabilities of SPACE, multiple datasets were used in the study. Download address:
Model architecture: A cell-cell interaction-aware cell-embedded model
SPACE uses a graph autoencoder framework to learn low-dimensional cell embeddings, which describe the gene expression information of each cell in the spatial transcriptome data and its interaction information with spatial neighboring cells (so the cell embedding is called cell-cell interaction-aware cell embedding). Based on this cell embedding, SPACE uses a clustering algorithm to identify spatial cell subtypes and discover tissue modules.
From the architecture point of view, the SPACE model consists of three parts: encoder (three-layer graph attention network), neighbor graph decoder and gene expression decoder. The following figure shows the overall framework of the model:
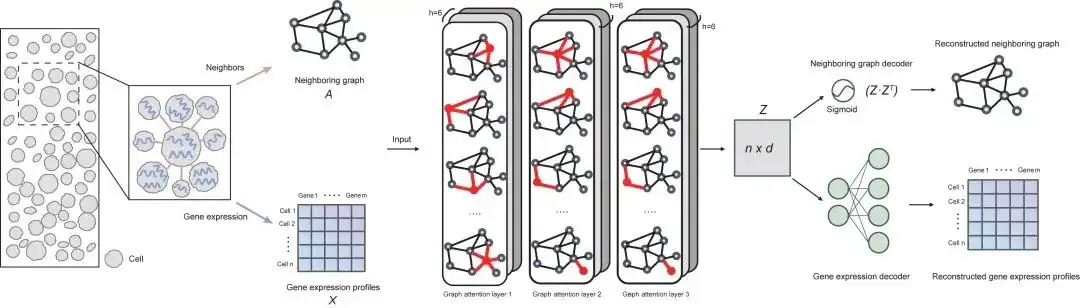
Performance Evaluation
* SPACE can identify biologically distinct cell types based on the spatial information in the ST dataset.
* SPACE outperforms currently available tools for distinguishing spatially informative cell types from ST data.
* SPACE outperforms state-of-the-art tools in tissue module discovery.
Tsinghua University Zhang Qiangfeng Laboratory

Zhang Qiangfeng's laboratory is affiliated with the School of Life Sciences of Tsinghua University. It is also an important part of the Tsinghua-Peking University Joint Center for Life Sciences and the Beijing Advanced Innovation Center for Structural Biology.
The laboratory's research focuses on interdisciplinary fields such as structural biology, genomics, machine learning and big data analysis. The main research direction is to combine structural biology and systems biology, develop and use a combination of computational and experimental methods to interpret the structure and function relationship of biological macromolecules (such as proteins, RNA, DNA), reconstruct their interaction networks, and discover the pathogenesis and possible treatments of complex diseases (including cancer and infectious diseases) related to changes in protein and RNA structure and abnormal macromolecular interactions.
The laboratory has unique protein and RNA structure modeling, RNA structure measurement based on next-generation sequencing, high-throughput RNA-protein interaction detection technology, and powerful computational and experimental platforms to advance researchers' cutting-edge research.
Meet AI4S Live Series
HyperAI (hyper.ai) is China's largest search engine in the field of data science. It focuses on the latest scientific research results of AI for Science and tracks academic papers in top journals such as Nature and Science in real time. So far, it has completed the interpretation of more than 100 AI for Science papers.
In addition, we also operate the only AI for Science open source project in China, awesome-ai4s.
* Project address:
https://github.com/hyperai/awesome-ai4s
In order to further promote the popularization of AI4S, further reduce the dissemination barriers of scientific research results of academic institutions, and share them with a wider range of industry scholars, technology enthusiasts and industrial units, HyperAI has planned the "Meet AI4S" video column, inviting researchers or related units who are deeply engaged in the field of AI for Science to share their research results and methods in the form of videos, and jointly discuss the opportunities and challenges faced by AI for Science in the process of scientific research progress and promotion and implementation, so as to promote the popularization and dissemination of AI for Science.