Command Palette
Search for a command to run...
New Uses for Old Drugs: AdaDR Released by the Central South University Team for Drug Repositioning Based on Adaptive Graph Convolutional Networks

In modern society, humans need to continue to fight against increasingly prevalent complex diseases such as tumors, diabetes, and cardiovascular diseases. Existing drugs can no longer fully meet market demand, and new drug research and development is imperative. However, the traditional drug discovery process is time-consuming and requires large investments. If we can proactively screen new drugs and therapeutic targets from past drugs and abandoned compounds, we can obviously significantly save R&D costs and improve R&D efficiency.
Drug repositioning, or “repurposing old drugs for new uses”, is an FDA-approved drug development approach that applies existing treatments to new disease processes. For example, sildenafil was originally used to treat chest pain, but was later found to be a PDE5 (phosphodiesterase type 5 inhibitor), which made sildenafil very popular in the market.
Due to its advantages such as reducing drug risks, shortening clinical evaluation cycles, low cost and high efficiency, repositioning of existing drugs has become a hot topic in current industry research.With the rapid development of deep learning, graph convolutional networks (GCNs) have been widely used in drug repositioning tasks. However, existing GCN-based methods have limitations in deeply integrating node features and topological structures. In response to this, researchers from Central South University published a paper titled "Drug repositioning with adaptive graph convolutional networks" in Bioinformatics.
This study proposed an adaptive GCN method called AdaDR to perform drug repositioning by deeply integrating node features and topological structures.Different from traditional graph convolutional networks, AdaDR simulates the interactive information between them through adaptive graph convolution operations, thereby enhancing the expressive power of the model.
Specifically, AdaDR extracts embeddings from both node features and topological structures, and uses an attention mechanism to learn adaptive importance weights of the embeddings.
Experimental results show that AdaDR outperforms several baseline methods in drug repositioning. In addition, exploratory analysis for discovering new drug-disease associations is provided in case studies.
Research highlights:
* This study proposes an adaptive graph convolutional network framework for drug relocalization tasks, performing graph convolution operations on topological structures and feature spaces.
* Considering the differences between topological structures and features, this study uses the attention mechanism to fully integrate them to distinguish their contributions to the model results
* The model proposed in this study is practical in drug repositioning tasks and helps reduce the risk of drug development failure
Paper address:
https://academic.oup.com/bioinformatics/article/40/1/btad748/7467059
Dataset download address:
Follow the official account and reply "relocation" to get the complete PDF
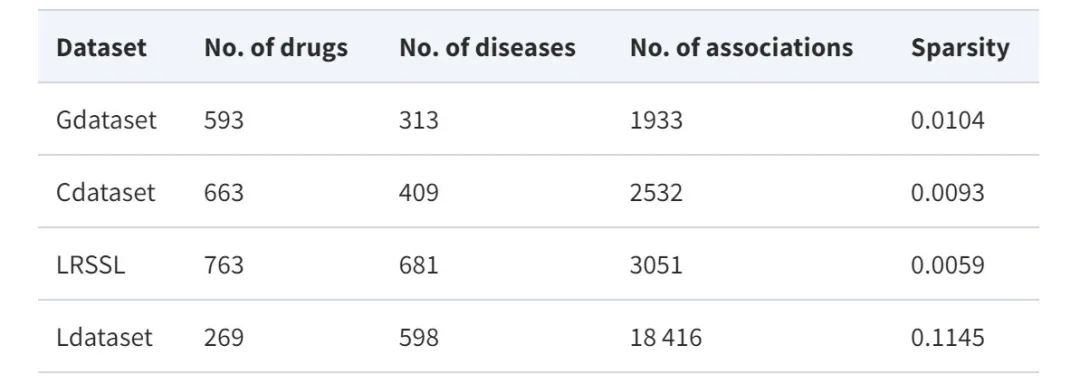
Dataset: Using four major benchmark datasets
In order to comprehensively evaluate the performance of the proposed model,This study utilized four benchmark datasets widely used in drug repositioning tasks, namely Gdataset, Cdataset, Ldataset and LRSSL.
* Gdataset:Considered the gold standard dataset, it includes 593 drugs from DrugBank and 1,933 confirmed drug-disease associations between 313 diseases listed in the OMIM database.
* Cdataset:Contains 663 drugs, 409 diseases, and 2,352 interacting drug-disease associations.
* Ldataset:Compiled from the CTD dataset, it includes 18,416 associations between 269 drugs and 598 diseases.
*LRSSL:A total of 3,051 validated drug-disease associations involving 763 drugs and 681 diseases were included.
At the same time, in order to construct the drug/disease feature map, the study also used the similarity characteristics of drugs and diseases. The data set statistics are shown in the following table:
Model architecture: A new adaptive GCNs framework AdaDR
The AdaDR model framework proposed in this study mainly consists of three components, as shown in the following figure:
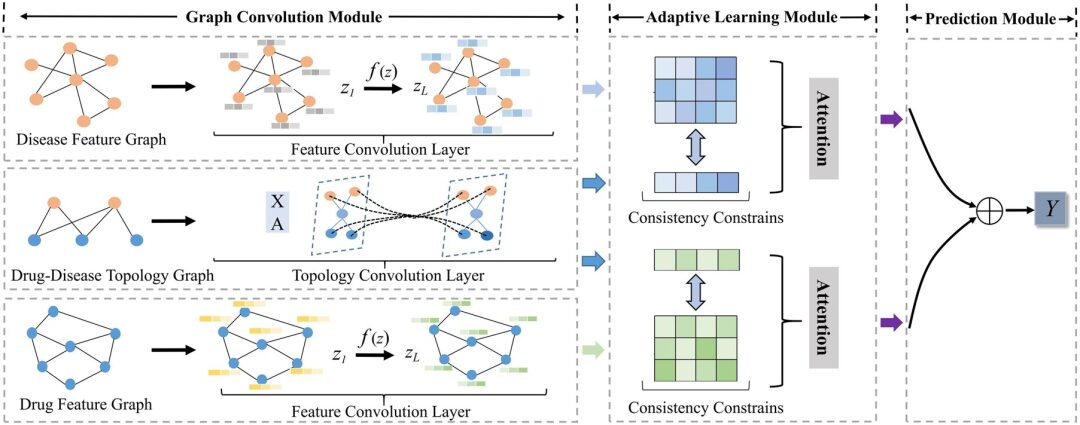
* Graph Convolution Module:Contains feature convolutional layers and topological convolutional layers to represent drug/disease embeddings in feature space and topological space.
* Adaptive Learning Module:Attention mechanism is used to distinguish the importance of the acquired embeddings. In this module, consistency constraint is used to extract common semantic information between feature and topological spaces.
* Prediction Module:The embeddings are concatenated together as the output to predict the result.
Research results: AdaDR outperforms multiple baseline methods in drug repositioning
Overall, AdaDR, as a new model, can significantly improve the performance of drug repositioning tasks.
First, the performance in cross validation:This study performed 10-fold cross-validation on AdaDR and other models, and calculated the mean and standard deviation of the results.
According to the results, due to AdaDR's feature integration capability, its final average results on four datasets obtained in 10 10-fold cross validations are better than all the compared methods.
For example, on the four benchmark datasets Gdataset, Cdataset, LRSSL and Ldataset,The results of this study are 9.8%, 9.1%, 9.1% and 7.1% higher than the AUPRC (area under the precision-recall) of the second-best method DRHGCN, respectively, fully demonstrating the effectiveness of the new method.
Next is the ability to predict the potential indications of new drugs:This study conducted a new experiment to evaluate the ability of AdaDR to predict the potential indications of new drugs.
Compared with the other 7 methods, AdaDR achieved the best performance (the blue bar in the figure below represents AdaDR). In terms of AUROC (area under the receiver operating characteristic curve), as shown in the figure below (a), AdaDR achieved an AUROC value of 0.948, which is better than other methods. At the same time, as shown in the figure below (b), AdaDR achieved an AUPRC of 0.393, which is higher than all other methods.
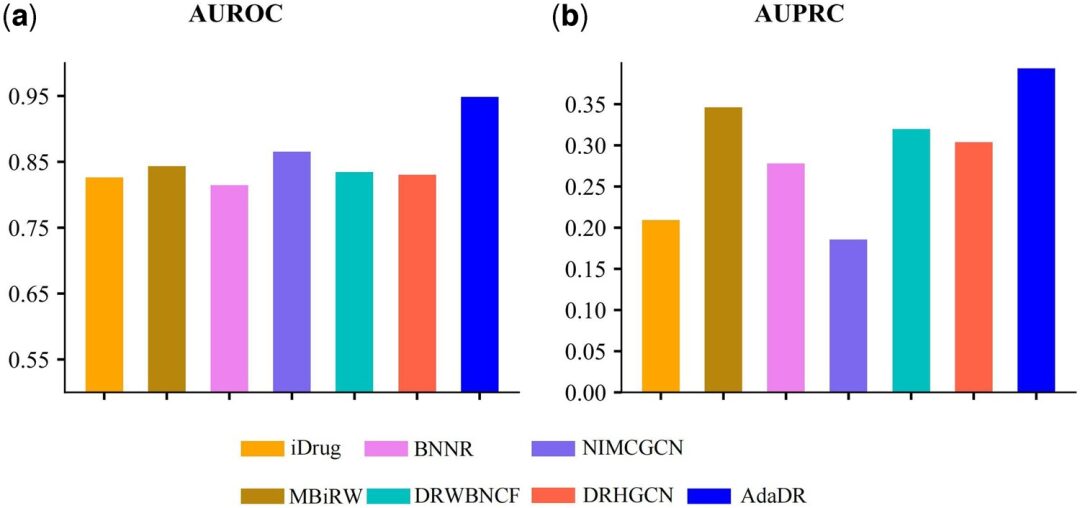
(a) AUROC of prediction results obtained by applying AdaDR and other competing methods.
(b) AUPRC of the prediction results obtained by applying AdaDR and other competing methods.
It is worth mentioning that in order to further verify the performance of AdaDR, the research team also applied AdaDR to predict candidate drugs for Alzheimer's disease (AD) and breast cancer (BRCA).
Among them, Alzheimer's disease is a gradually developing neurodegenerative disease for which there is currently no effective drug. Breast cancer is a phenomenon in which breast epithelial cells proliferate uncontrollably under the influence of multiple carcinogenic factors. Although there are currently a variety of drugs for the treatment of breast cancer, such as paclitaxel and carboplatin, more drug options may provide better treatment options. The following table reports candidate drugs with evidence support:
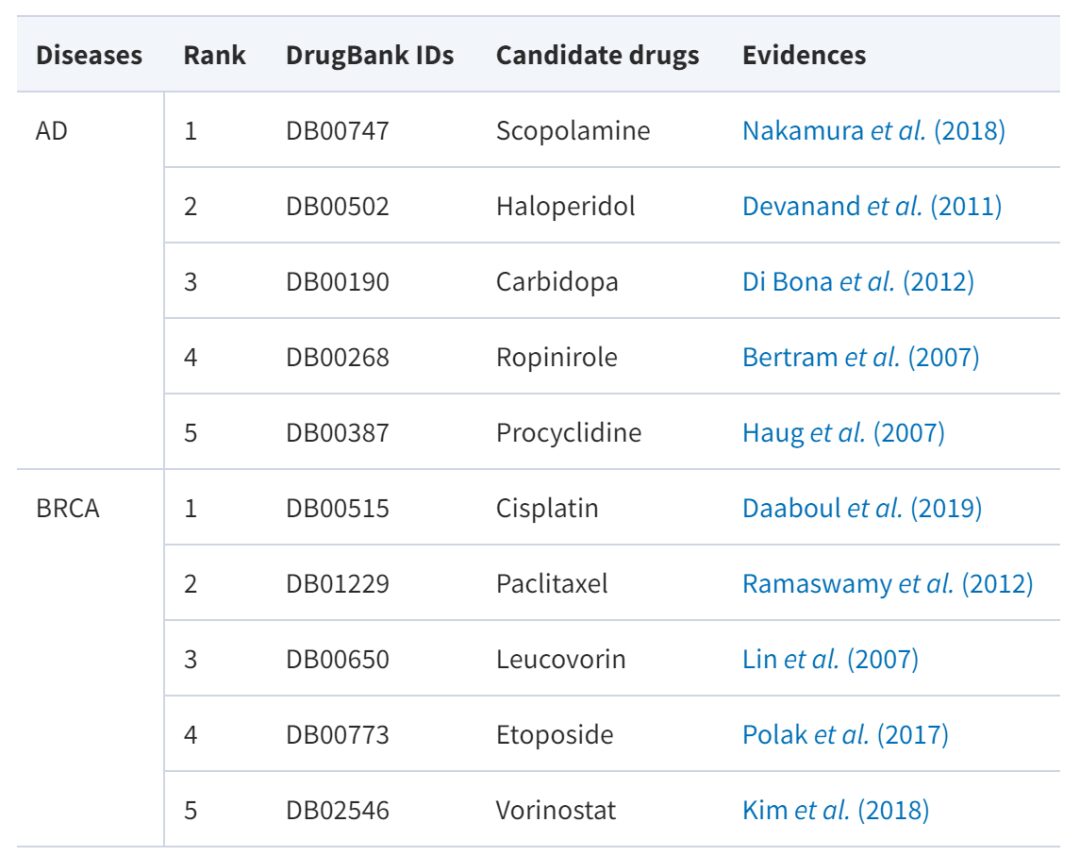
It can be seen that among the top five drugs in AdaDR prediction scores, many have been verified by authoritative sources and literature (success rate 100%). In addition, the model of this study can produce interpretable results. Taking paclitaxel as an example, the model predicts that it can treat breast cancer. This is indeed supported by authoritative sources and literature.
Interestingly, the researchers found that docetaxel appeared in their training set. Paclitaxel and docetaxel are similar molecules with the same paclitaxel core.This reflects that the new model can utilize drug similarity information to make meaningful predictions.
The return on investment in pharmaceutical R&D continues to decline, and drug repositioning may be the key to breaking the deadlock
Today, pharmaceutical companies are in the midst of unprecedented change. The COVID-19 pandemic and the ensuing economic recession have left pharmaceutical companies facing a series of challenges and uncertainties, and the return on innovation has become a top priority for every pharmaceutical company.
Although biopharmaceutical companies have invested heavily in R&D for innovation over the past 10 years, the return rate has dropped significantly over the same period. The "2019 Pharmaceutical Innovation Return Rate Assessment" released by the Deloitte Centre for Health Solutions shows that the return on investment in pharmaceutical R&D in 2019 was the lowest since 2010, at only 1.8%. According to the data shown in the ten reports, the return on investment of pharmaceutical companies' R&D has been on a downward trend over the past decade.
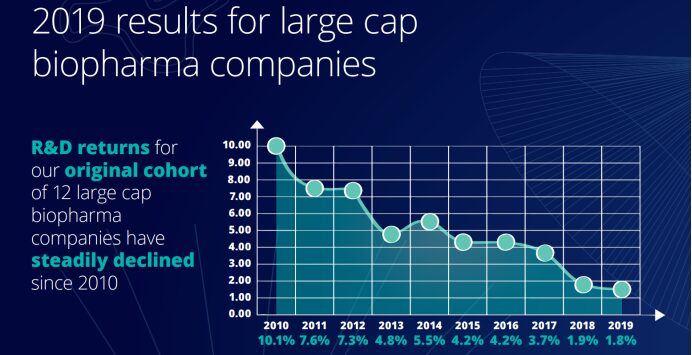
Not only that, the peak sales of each new drug after its launch also fell from $407 million in 2018 to $376 million in 2019, falling below $400 million for the first time and less than half of the $816 million in 2010. Compared with 2010, the cost of launching a new drug increased by 67%, from $1.188 billion in 2010 to $1.981 billion in 2019.The decline in peak sales stands in stark contrast to the increase in the average cost of bringing a new drug to market, suggesting that pharmaceutical companies are spending longer than ever before in the research and development process.
Repositioning drugs can save the early costs and time required to bring drugs to market, thereby accelerating the transition from basic research to clinical treatment. According to industry insiders, a new drug must go through a series of studies in vitro, preclinical animals, clinical phases I, II, and III from the beginning of research and development to approval for marketing. It takes 10 to 15 years and costs at least $1 billion. In contrast, some survey results show that the average cost of repositioning drugs is only $300 million, and it takes about 6.5 years to enter the market.
Drug repositioning mainly includes methods based on machine learning, methods based on big data mining and positioning, and methods based on in vivo positioning.Compared with in vivo methods, drug repositioning technology based on machine learning and big data mining has the advantages of fast speed and low cost, and has become a potentially powerful technology.
The article "A Review of Drug Repositioning Algorithms Based on Machine Learning and Big Data Mining" introduces the research progress of computational drug repositioning in recent years.
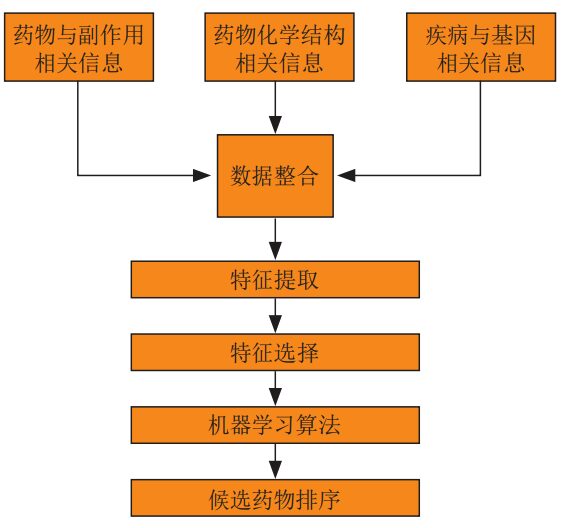
in,Based on the traditional machine learning algorithm method,First, the drug and side effect information, drug chemical structure information, and disease and gene related information are integrated, and then the training data is obtained through feature extraction and feature selection, and then the relevant machine learning algorithm is selected for training, and finally the trained algorithm model is used to obtain the drug repositioning results.
In deep learning based methods,Some researchers have systematically compared deep neural networks with a variety of other machine learning methods in multiple aspects of drug development. The results show that deep learning performs better than traditional machine learning algorithms.
In the network similarity reasoning method,A research team from East China University of Science and Technology proposed a network-based inference (NBI) method that infers new targets of known drugs using only the topological similarity of drug-target bipartite networks.
With the development of big data mining technology, drug repositioning based on machine learning and big data mining algorithms will provide more effective methods for the treatment of diseases and has become the focus of biomedical research. There is reason to believe that rational reasoning and computational models will play an important role in the future drug repositioning process.
References:
1.https://www.cn-healthcare.com/article/20191224/content-527902.html
2.https://pps.cpu.edu.cn/cn/article/pdf/preview/b286f85e-a37a-4007-ab94-918629aef556.pdf
3.https://mp.weixin.qq.com/s/lD-HyfwUHiX4f-llS6lykQ