Command Palette
Search for a command to run...
Learn About the PDB Protein Structure Dataset Behind AlphaFold 2 in One Article
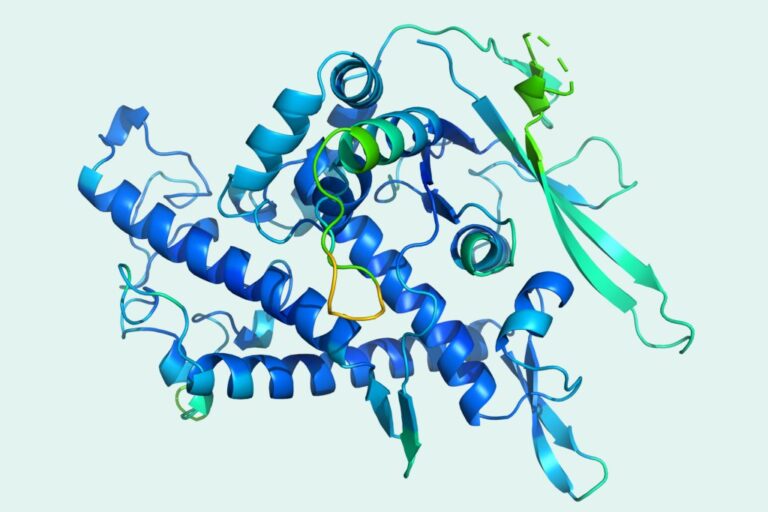
DeepMind's latest generation algorithm, AlphaFold 2, has completely defeated other competitors in CASP, which is recently known as the "Protein Olympics", and has achieved amazing breakthroughs, shocking the entire scientific research community. After being overwhelmed by this scientific research achievement, let's take a look at the data set behind the algorithm.
In the past two days, we have been bombarded with news about DeepMind's new generation artificial intelligence algorithm AlphaFold 2, especially in the biological world, which can be said to have ushered in a milestone breakthrough.
According to DeepMind's official announcement, its deep learning algorithm AlphaFold 2 has successfully solved major problems in the biological field over the past 50 years.
The algorithm can accurately predict the 3D structure of proteins based on their amino acid sequences, with an accuracy comparable to 3D structures solved using experimental techniques such as cryo-electron microscopy (CryoEM), nuclear magnetic resonance or X-ray crystallography.
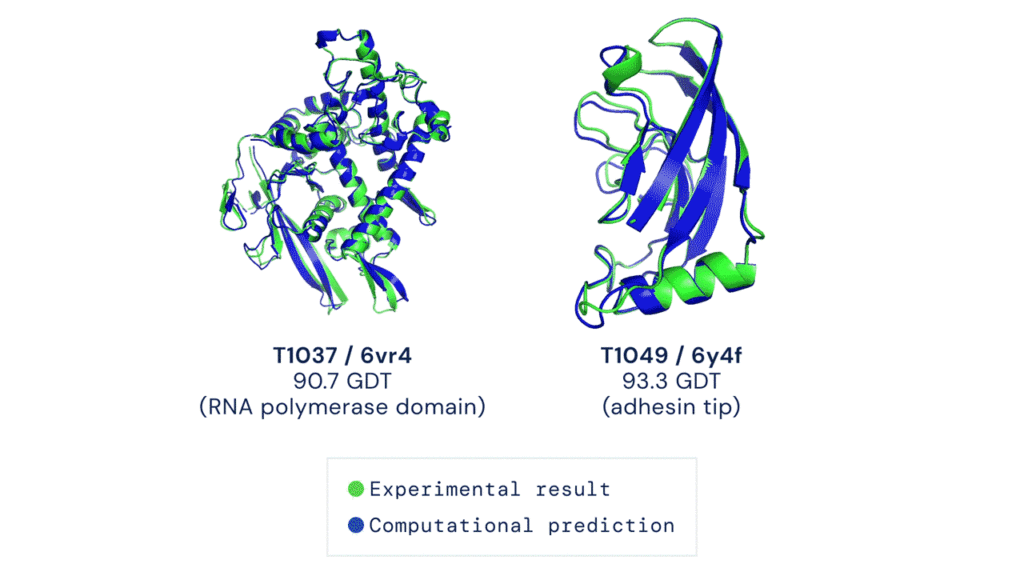
This milestone event excited biologists, but it also made many people in the industry tremble with fear, and they said they would switch careers to study deep learning.
However, while everyone is paying attention to this scientific research result, don’t forget the hero behind it—— PDB protein structure data set, a data set dedicated to collecting three-dimensional structure data of proteins and nucleic acids.
This data set is essential for a groundbreaking breakthrough
According to DeepMind, the team trained the system on public data.These data come from the protein structure dataset PDB and the large database UniProt containing protein sequences of unknown structures, which together include about 170,000 protein structures.
in,PDB is a data set dedicated to the three-dimensional structure of proteins and nucleic acids. It has a very long history, dating back to 1971.
Walter Hamilton of Brookhaven National Laboratory in the United States decided to establish this database. In October 1998, PDB was handed over to the Research Collaboratory for Structural Bioinformatics (RCSB), which was managed by Helen M. Berman of Rutgers University, who was also a member of RCSB.

In 2003,The PDB evolved into an international organization, wwPDB (Worldwide Protein Database), to oversee the PDB resources. Other members of wwPDB, including PDBe (Europe), RCSB (USA), and PDBj (Japan), also provide centers for data accumulation, processing, and publication for PDB.

It is worth mentioning that although the data in PDB are submitted by scientists from all over the world, each submitted data will be reviewed and annotated by wwPDB staff to check whether the data is reasonable. PDB and the software it provides are now open to the public free of charge.
More than 140,000 structures, what information is in the PDB?
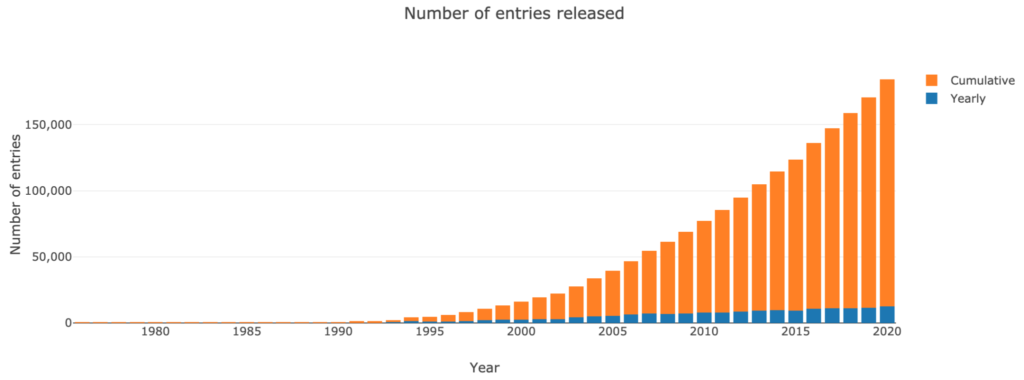
Over the past few decades, the number of structures in the PDB has grown at a near exponential rate:
- 100 in 1982;
- 1,000 in 1993;
- 10,000 in 1999;
- 100,000 in 2014.
However, since 2007, the rate at which new protein structures are accumulated appears to have stabilized.
Structural biologists around the world use methods such as X-ray crystallography, NMR spectroscopy and cryo-electron microscopy to determine the position of each atom in a molecule relative to each other. They then submit this structural information, which is annotated and publicly released to the wwPDB.
You can search for structures of ribosomes, oncogenes, drug targets, and even entire viruses in the PDB dataset.However, the sheer number of structures archived in the PDB makes finding the information you need potentially a daunting task.
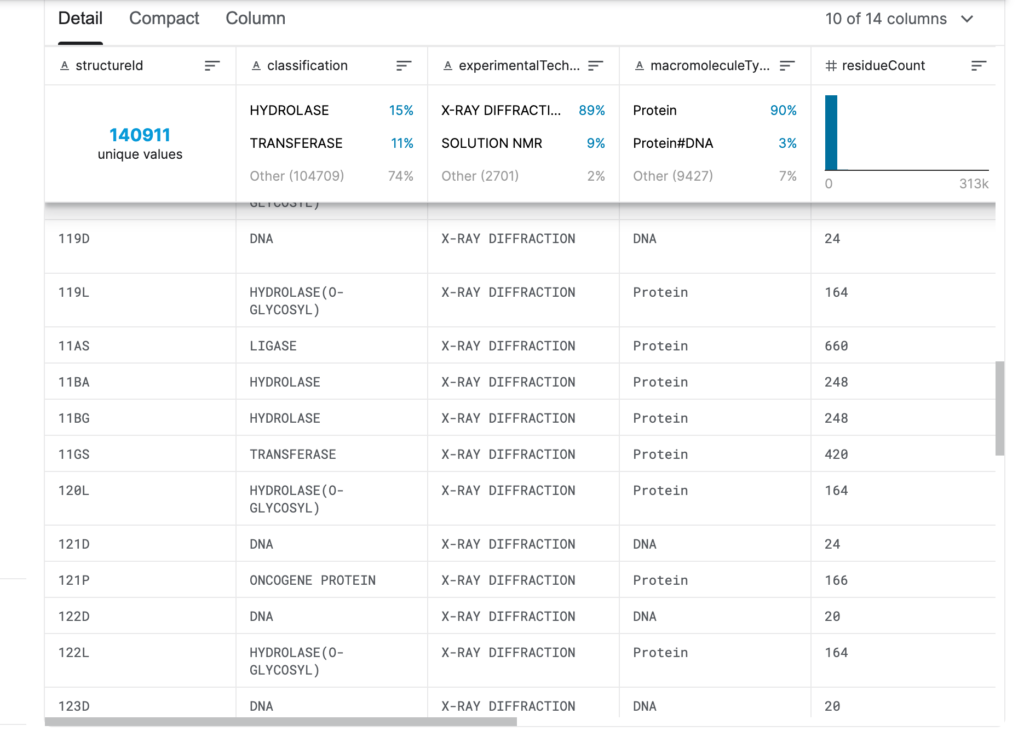
The information in the PDB dataset mainly includes:The source of protein/nucleic acid, the composition of protein/nucleic acid molecules, atomic coordinates, experimental methods used to determine the structure, as well as other data and information such as temperature factors and structure determiners.
How to download?
The dataset is now available on the official website of Hyperneuron and openbayes.com. Visit:https://orion.hyper.ai/datasets/13906 Or click "Read original text" to get the data set with one click.
■ Details of PDB protein structure dataset
Release time:Collected since 1971
Publishing Agency:wwPDB
Quantity included:140,000+ protein/nucleic acid structures
Data format:csv file
Data size:27 MB (146 MB after decompression)
Download address:https://orion.hyper.ai/datasets/13906
The same dataset as DeepMind, you deserve it too~
How to use?
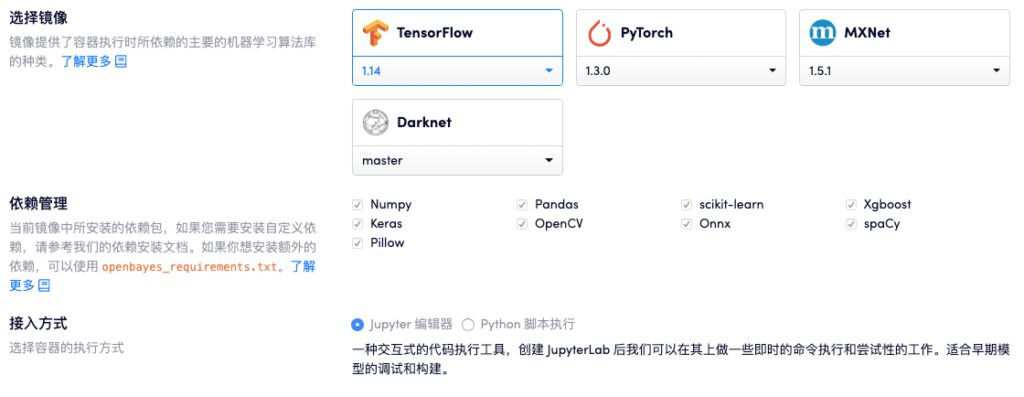
Our partner OpenBayes is a cloud service that provides cloud computing power for machine learning. They have a large-scale supercomputing cluster, and the GPU cluster architecture is designed specifically for matrix computing. It provides computing power containers for AI applications, and it is very easy to use and can be used out of the box.
Currently, OpenBayes' computing power container products already support TensorFlow, PyTorch, MXNet and other CPU and GPU environments, different versions and types of standard machine learning frameworks and various common dependencies.
Currently, the OpenBayes computing container supports standard librariesand provide CPU, NVIDIA T4, NVIDIA Tesla V100 and other computing resourcesWhether it is centralized training of massive data or low-power model resident operation, it can easily meet user needs.

From CPU to T4 to V100, a wide range of computing container configurations OpenBayes supportScript upload and JupyterLab editorOnline programming and then model training.
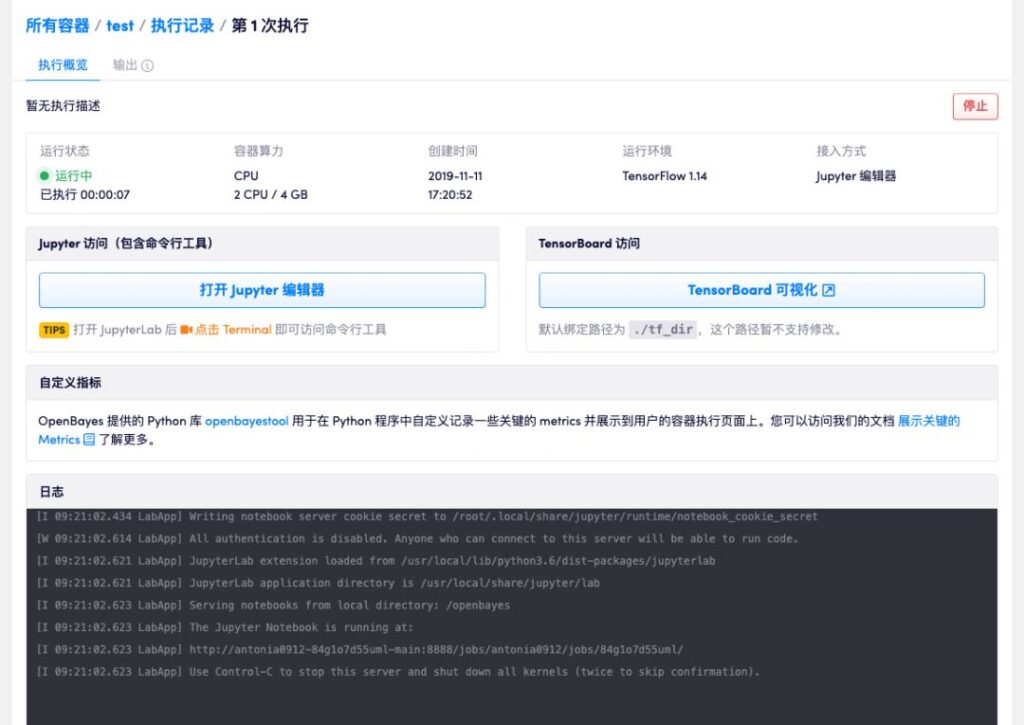
Clear and concise execution processFull tutorial: https://openbayes.com/docs/quickstart/
Register as a new user to enjoy GPU computing power
Visit openbayes.com, click on the official website to register immediately, and there will be free gifts every week during the internal test period, so you don’t have to compete with classmates and colleagues for computing power~
Event Description Visit openbayes.com Register as a new user with the invitation code [HyperAI]You can enjoy
Free CPU quota:300 minutes/week
Free vGPU quota:180 minutes/week
PDB full dataset acquisition:
https://www.rcsb.org/#Category-download
The files in the PDB dataset can be viewed directly with a text editor, but it is better to use a visualization tool. The officially recommended viewing program is Swiss PDB viewer:
https://spdbv.vital-it.ch/disclaim.html#
Other references:
https://www.novopro.cn/articles/201912021193.html
-- over--