Command Palette
Search for a command to run...
GROMACS 入门教程:水中的溶菌酶
以「水中的溶菌酶」为例进行分子动力学模拟
教程简介
该教程为使用 GROMACS 软件进行分子动力学模拟的一个入门教程,本次教程以 水中的溶菌酶 为例学习如何准备和运行一个典型的蛋白质在水中的分子动力学模拟。
在 OpenBayes 平台使用 NVIDIA RTX 4090 显卡配置的 GPU 版本的 GROMACS,经过 GPU 并行计算后的计算效率大大提高,速度可达到 255ns/day!以下是速度表现:
Core t (s) Wall t (s) (%)
Time: 3972.923 198.659 1999.9
(ns/day) (hour/ns)
Performance: 255.471 0.094
GROMACS 简介
GROMACS (GROningen MAchine for Chemical Simulations) 是一个用于分子动力学模拟的高性能软件包,主要应用于对生物分子(如蛋白质、脂质和核酸)在不同条件下的运动行为进行建模和模拟。它最初由荷兰格罗宁根大学开发,目前已成为分子动力学领域中最常用的开源工具之一。
GROMACS 的主要特点
1. 高性能:
• GROMACS 对并行计算进行了高度优化,可以在现代多核 CPU 和 GPU 系统上高效运行。
• 支持 OpenMP 和 MPI 以实现多线程和分布式计算。
2. 广泛的应用范围:
• 可用于模拟从小分子到大型蛋白质复合物的行为。
• 支持生物分子、聚合物、无机化合物以及其他化学体系的研究。
3. 灵活性:
• 提供了丰富的工具用于前处理(如拓扑生成、溶剂化)和后处理(如轨迹分析、能量计算)。
• 支持多种力场,如 AMBER 、 CHARMM 和 GROMOS 。
4. 易用性:
• GROMACS 包含用户友好的命令行界面。
• 提供了详细的文档和教程,适合初学者和高级用户。
5. 开源和可扩展性:
• 开源许可证允许用户修改和扩展 GROMACS 以适应特定需求。
• 拥有活跃的用户社区和开发团队。
GROMACS 一般使用流程
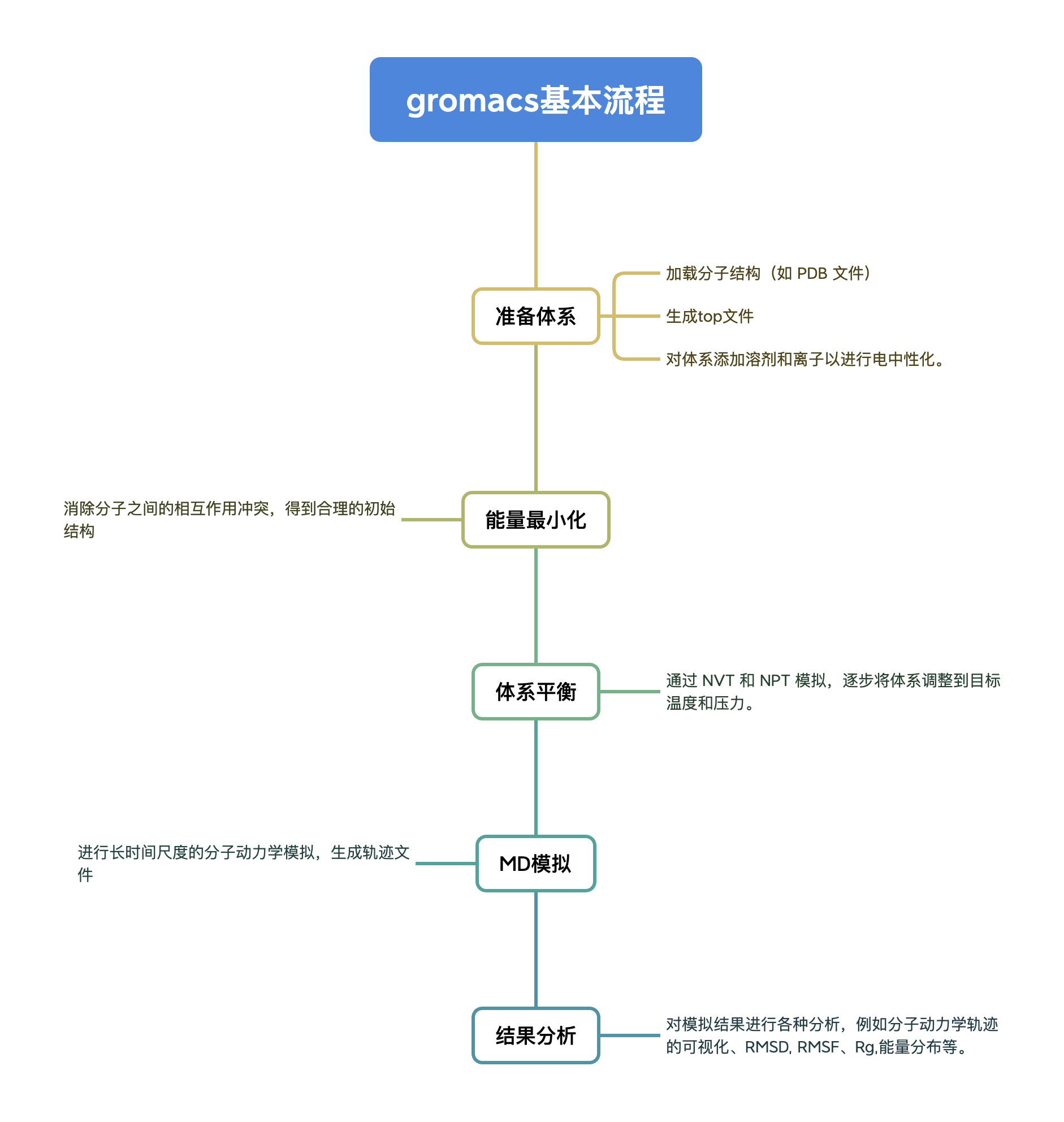
运行步骤
首先我们需要配置 pdb 蛋白文件,md 计算文件,再经过各种预处理后,最终进行了 10ns 模拟,并对结果进行分析。以下为分步详解。
1. 启动 GROMACS
首先登录平台:https://openbayes.com/
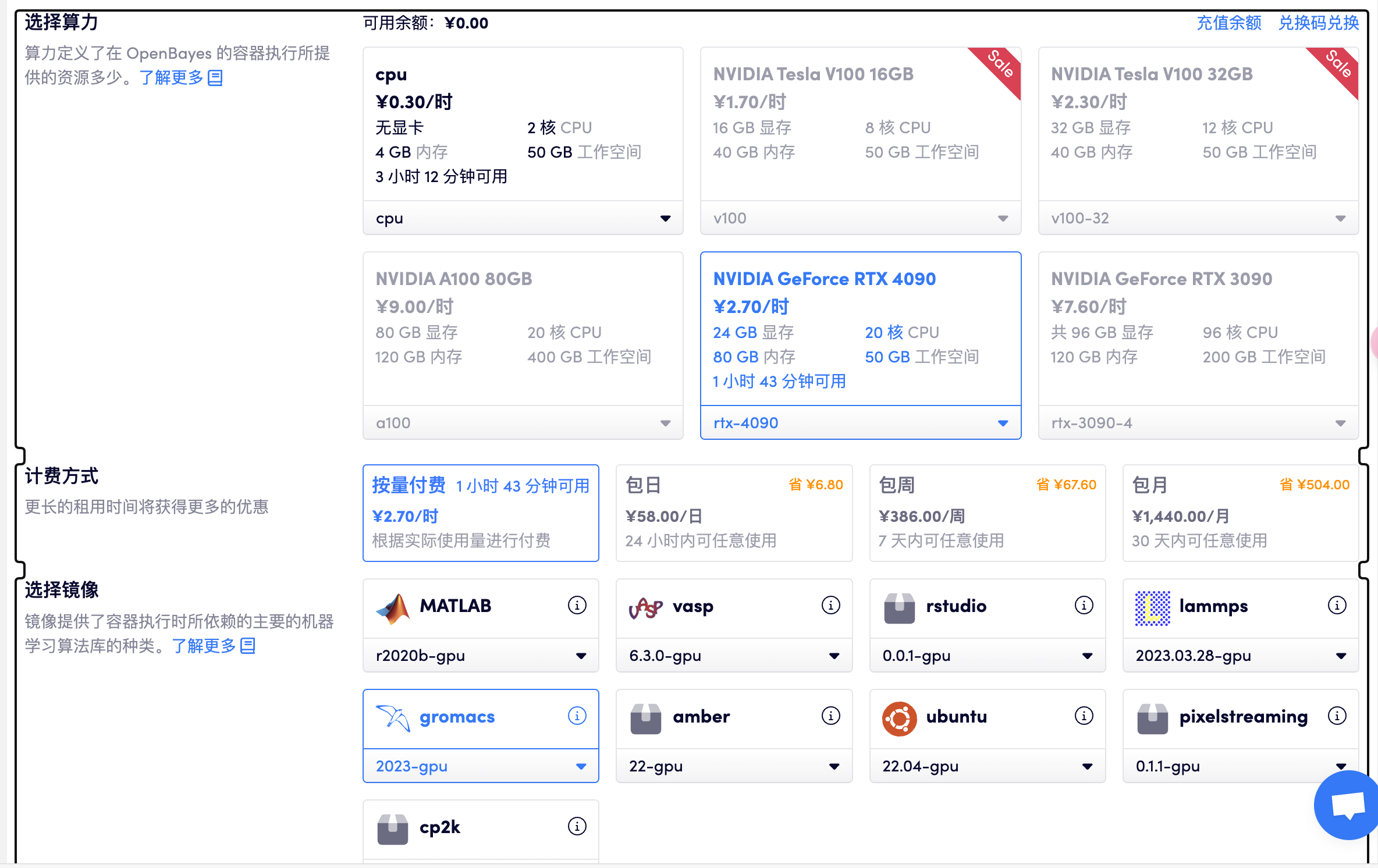
选择「高性能计算」> 创建新容器> 选择算力 RTX 4090> 选择 gromacs GPU
打开工作空间
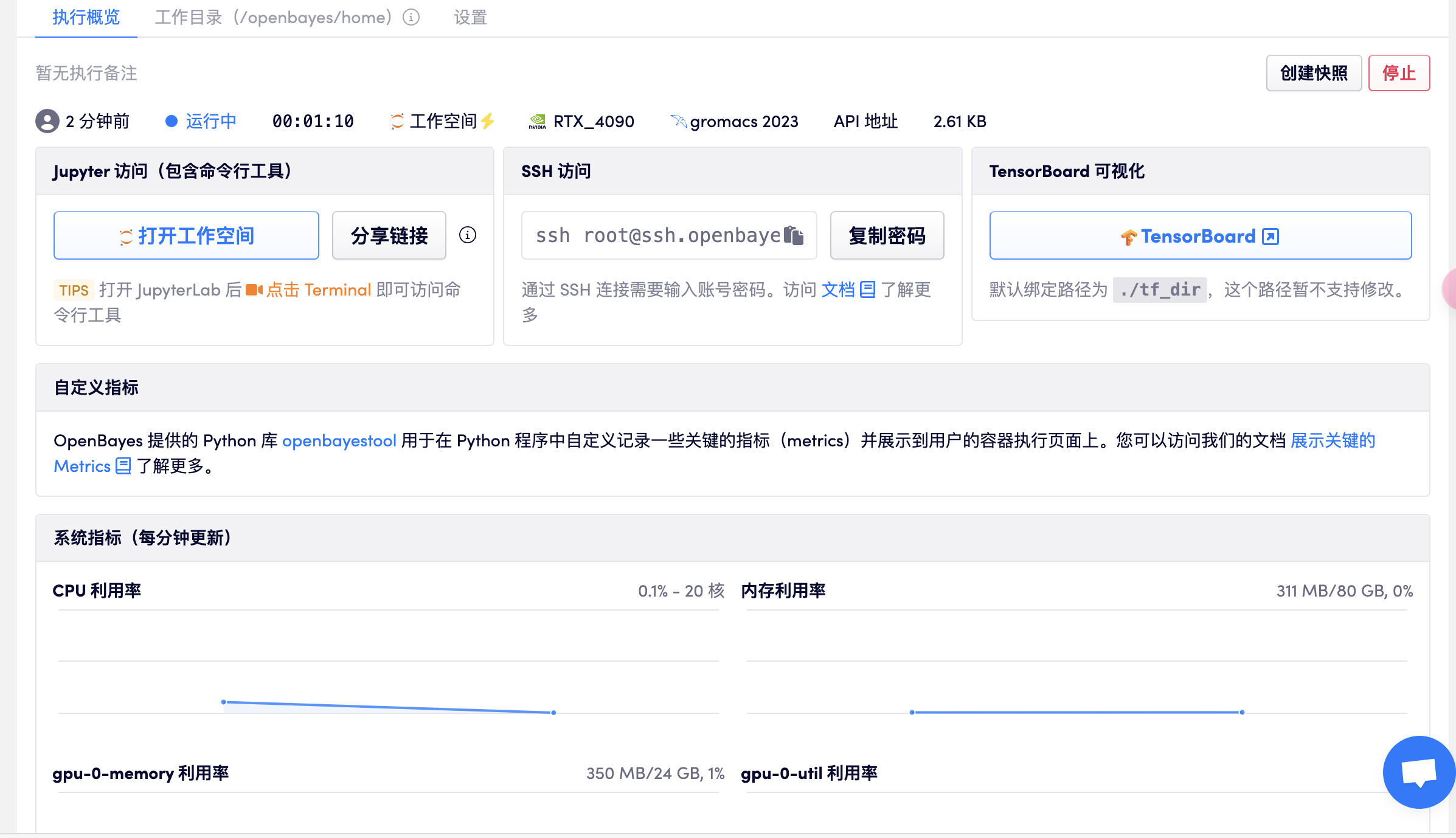
打开终端
或者采用 SSH 控制服务器:
在 X-shell 或者 mac unix 终端,输入:ssh -p 32699 [email protected],再输入密码即可(如下所示)↓
liangzhongzhongzhong@lzr ~ % ssh -p 32699 [email protected]
The authenticity of host '[ssh.openbayes.com]:32699 ([101.237.34.75]:32699)' can't be established.
ED25519 key fingerprint is SHA256:uwPyhP/EYoW49Ez4rvAuaf19czwis2rdS4pImsR0NH8.
This key is not known by any other names.
Are you sure you want to continue connecting (yes/no/[fingerprint])? yes
Warning: Permanently added '[ssh.openbayes.com]:32699' (ED25519) to the list of known hosts.
[email protected]'s password:
OpenBayes
目录说明
- /openbayes/home 工作空间内的数据保存地址,容器停止后,该目录中的内容不会被删除
- /openbayes/input/input0 - /openbayes/input/input4 为数据目录,不会占用工作空间的存储容量,最多支持同时绑定 5 个
⚠️ 其他目录下的内容在容器关闭后会被自动删除!更多信息请访问 https://openbayes.com/docs/concepts
⚠️ 禁止挖矿,一经发现将立即封号恕不退款
(base) root@liangzhong-4ay9ej85pxvd-main:/openbayes/home# ls
(base) root@liangzhong-4ay9ej85pxvd-main:/openbayes# l(base) root@liangzhong-4ay9ej85pxvd-main:/openbayes# lhome input 请将文件存在 home 目录下, 当前文件夹下的文件不会被保存.txt
(base) root@liangzhong-4ay9ej85pxvd-main:/openbayes# ls(base) root@liangzhong-4ay9ej85pxvd-main:/openbaye
(base) root@liangzhong-4ay9ej85pxvd-main:/openbayes/input# cd input0
(base) root@liangzhong-4ay9ej85pxvd-main:/openbayes/input/input0# ls
'#nvt.log.1#' '#topol.top.2#' 1AKI_processed.gro 1aki.pdb em.log ions.mdp mdout.mdp nvt.cpt nvt.log nvt.trr topol.top
'#nvt.log.2#' 1AKI_clean.pdb 1AKI_solv.gro em.edr em.tpr ions.tpr minim.mdp nvt.edr nvt.mdp posre.itp
'#topol.top.1#' 1AKI_newbox.gro 1AKI_solv_ions.gro em.gro em.trr md.mdp npt.mdp nvt.gro nvt.tpr potential.xvg
(base) root@liangzhong-4ay9ej85pxvd-main:/openbayes/input/input0#

调用 GROMACS,设置临时环境变量
export PATH=/data/app/gromacs/bin:$PATH
(base) root@liangzhong-4ay9ej85pxvd-main:/openbayes/home# gmx_mpi -h
:-) GROMACS - gmx_mpi, 2023 (-:
Executable: /data/app/gromacs/bin/gmx_mpi
Data prefix: /data/app/gromacs
Working dir: /output
Command line:
gmx_mpi -h
SYNOPSIS
gmx [-[no]h] [-[no]quiet] [-[no]version] [-[no]copyright] [-nice <int>]
[-[no]backup]
OPTIONS
Other options:
-[no]h (no)
Print help and quit
-[no]quiet (no)
Do not print common startup info or quotes
-[no]version (no)
Print extended version information and quit
-[no]copyright (no)
Print copyright information on startup
-nice <int> (19)
Set the nicelevel (default depends on command)
-[no]backup (yes)
Write backups if output files exist
Additional help is available on the following topics:
commands List of available commands
selections Selection syntax and usage
To access the help, use 'gmx help <topic>'.
For help on a command, use 'gmx help <command>'.
GROMACS reminds you: "All You Need is Greed" (Aztec Camera)
2. 文件准备
在运行教程前,我们需要准备以下 6 个文件: 1aki.pdb 、 ions.mdp 、 md.mdp 、 minim.mdp 、 npt.mdp 、 nvt.mdp
这些文件可以直接下载,或者按照下面的方式去创建。
例如 1AKI.pdb 蛋白文件 :可以从 RCSB 网站获取 1AKI.pdb 。教程中已将其他文件用 vim 文本编辑器,把代码复制过来创建。
RCSB 数据库介绍:
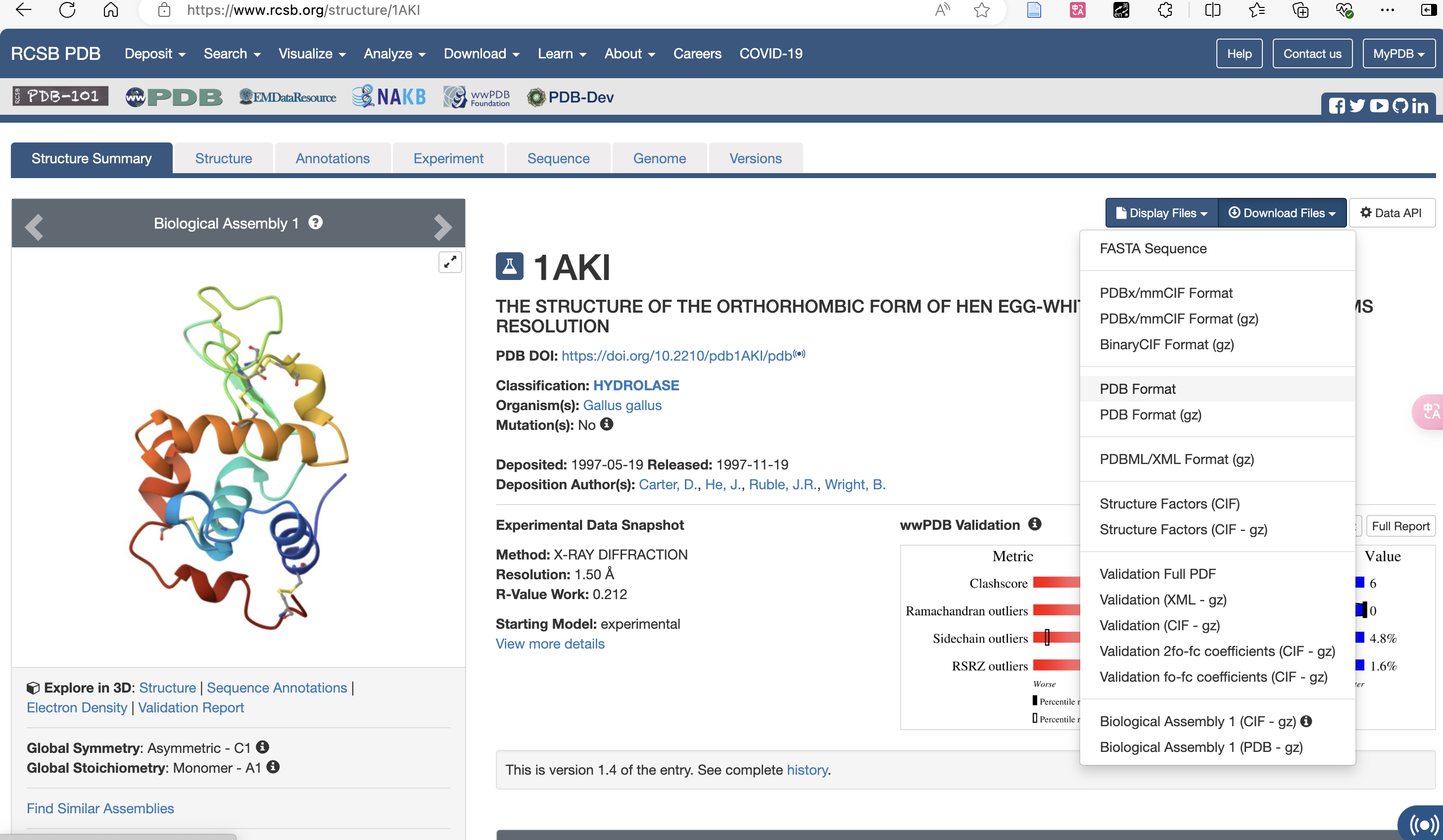
下载 pdb Format 格式:
上传到你的工作目录
(base) root@liangzhong-4ay9ej85pxvd-main:/input0# ls
1aki.pdb
#查看上传成功
vim ions.mdp
#准备 mdp 文件,输入命令后复制一下内容,按 i 进入插入模式开始编辑。
#编辑完成后,按 Esc 键退出插入模式,输入 :wq 保存并退出。
; ions.mdp - used as input into grompp to generate ions.tpr
; Parameters describing what to do, when to stop and what to save
integrator = steep ; Algorithm (steep = steepest descent minimization)
emtol = 1000.0 ; Stop minimization when the maximum force < 1000.0 kJ/mol/nm
emstep = 0.01 ; Minimization step size
nsteps = 50000 ; Maximum number of (minimization) steps to perform
; Parameters describing how to find the neighbors of each atom and how to calculate the interactions
nstlist = 1 ; Frequency to update the neighbor list and long range forces
cutoff-scheme = Verlet ; Buffered neighbor searching
ns_type = grid ; Method to determine neighbor list (simple, grid)
coulombtype = cutoff ; Treatment of long range electrostatic interactions
rcoulomb = 1.0 ; Short-range electrostatic cut-off
rvdw = 1.0 ; Short-range Van der Waals cut-off
pbc = xyz ; Periodic Boundary Conditions in all 3 dimensions
vim minim.mdp
; minim.mdp - used as input into grompp to generate em.tpr
; Parameters describing what to do, when to stop and what to save
integrator = steep ; Algorithm (steep = steepest descent minimization)
emtol = 1000.0 ; Stop minimization when the maximum force < 1000.0 kJ/mol/nm
emstep = 0.01 ; Minimization step size
nsteps = 50000 ; Maximum number of (minimization) steps to perform
; Parameters describing how to find the neighbors of each atom and how to calculate the interactions
nstlist = 1 ; Frequency to update the neighbor list and long range forces
cutoff-scheme = Verlet ; Buffered neighbor searching
ns_type = grid ; Method to determine neighbor list (simple, grid)
coulombtype = PME ; Treatment of long range electrostatic interactions
rcoulomb = 1.0 ; Short-range electrostatic cut-off
rvdw = 1.0 ; Short-range Van der Waals cut-off
pbc = xyz ; Periodic Boundary Conditions in all 3 dimensions
vim nvt.mdp
title = OPLS Lysozyme NVT equilibration
define = -DPOSRES ; position restrain the protein
; Run parameters
integrator = md ; leap-frog integrator
nsteps = 50000 ; 2 * 50000 = 100 ps
dt = 0.002 ; 2 fs
; Output control
nstxout = 500 ; save coordinates every 1.0 ps
nstvout = 500 ; save velocities every 1.0 ps
nstenergy = 500 ; save energies every 1.0 ps
nstlog = 500 ; update log file every 1.0 ps
; Bond parameters
continuation = no ; first dynamics run
constraint_algorithm = lincs ; holonomic constraints
constraints = h-bonds ; bonds involving H are constrained
lincs_iter = 1 ; accuracy of LINCS
lincs_order = 4 ; also related to accuracy
; Nonbonded settings
cutoff-scheme = Verlet ; Buffered neighbor searching
ns_type = grid ; search neighboring grid cells
nstlist = 10 ; 20 fs, largely irrelevant with Verlet
rcoulomb = 1.0 ; short-range electrostatic cutoff (in nm)
rvdw = 1.0 ; short-range van der Waals cutoff (in nm)
DispCorr = EnerPres ; account for cut-off vdW scheme
; Electrostatics
coulombtype = PME ; Particle Mesh Ewald for long-range electrostatics
pme_order = 4 ; cubic interpolation
fourierspacing = 0.16 ; grid spacing for FFT
; Temperature coupling is on
tcoupl = V-rescale ; modified Berendsen thermostat
tc-grps = Protein Non-Protein ; two coupling groups - more accurate
tau_t = 0.1 0.1 ; time constant, in ps
ref_t = 300 300 ; reference temperature, one for each group, in K
; Pressure coupling is off
pcoupl = no ; no pressure coupling in NVT
; Periodic boundary conditions
pbc = xyz ; 3-D PBC
; Velocity generation
gen_vel = yes ; assign velocities from Maxwell distribution
gen_temp = 300 ; temperature for Maxwell distribution
gen_seed = -1 ; generate a random seed
vim npt.mdp
title = OPLS Lysozyme NPT equilibration
define = -DPOSRES ; position restrain the protein
; Run parameters
integrator = md ; leap-frog integrator
nsteps = 50000 ; 2 * 50000 = 100 ps
dt = 0.002 ; 2 fs
; Output control
nstxout = 500 ; save coordinates every 1.0 ps
nstvout = 500 ; save velocities every 1.0 ps
nstenergy = 500 ; save energies every 1.0 ps
nstlog = 500 ; update log file every 1.0 ps
; Bond parameters
continuation = yes ; Restarting after NVT
constraint_algorithm = lincs ; holonomic constraints
constraints = h-bonds ; bonds involving H are constrained
lincs_iter = 1 ; accuracy of LINCS
lincs_order = 4 ; also related to accuracy
; Nonbonded settings
cutoff-scheme = Verlet ; Buffered neighbor searching
ns_type = grid ; search neighboring grid cells
nstlist = 10 ; 20 fs, largely irrelevant with Verlet scheme
rcoulomb = 1.0 ; short-range electrostatic cutoff (in nm)
rvdw = 1.0 ; short-range van der Waals cutoff (in nm)
DispCorr = EnerPres ; account for cut-off vdW scheme
; Electrostatics
coulombtype = PME ; Particle Mesh Ewald for long-range electrostatics
pme_order = 4 ; cubic interpolation
fourierspacing = 0.16 ; grid spacing for FFT
; Temperature coupling is on
tcoupl = V-rescale ; modified Berendsen thermostat
tc-grps = Protein Non-Protein ; two coupling groups - more accurate
tau_t = 0.1 0.1 ; time constant, in ps
ref_t = 300 300 ; reference temperature, one for each group, in K
; Pressure coupling is on
pcoupl = Parrinello-Rahman ; Pressure coupling on in NPT
pcoupltype = isotropic ; uniform scaling of box vectors
tau_p = 2.0 ; time constant, in ps
ref_p = 1.0 ; reference pressure, in bar
compressibility = 4.5e-5 ; isothermal compressibility of water, bar^-1
refcoord_scaling = com
; Periodic boundary conditions
pbc = xyz ; 3-D PBC
; Velocity generation
gen_vel = no ; Velocity generation is off
vim md.md
title = OPLS Lysozyme NPT equilibration
; Run parameters
integrator = md ; leap-frog integrator
nsteps = 500000 ; 2 * 500000 = 1000 ps (1 ns)
dt = 0.002 ; 2 fs
; Output control
nstxout = 0 ; suppress bulky .trr file by specifying
nstvout = 0 ; 0 for output frequency of nstxout,
nstfout = 0 ; nstvout, and nstfout
nstenergy = 5000 ; save energies every 10.0 ps
nstlog = 5000 ; update log file every 10.0 ps
nstxout-compressed = 5000 ; save compressed coordinates every 10.0 ps
compressed-x-grps = System ; save the whole system
; Bond parameters
continuation = yes ; Restarting after NPT
constraint_algorithm = lincs ; holonomic constraints
constraints = h-bonds ; bonds involving H are constrained
lincs_iter = 1 ; accuracy of LINCS
lincs_order = 4 ; also related to accuracy
; Neighborsearching
cutoff-scheme = Verlet ; Buffered neighbor searching
ns_type = grid ; search neighboring grid cells
nstlist = 10 ; 20 fs, largely irrelevant with Verlet scheme
rcoulomb = 1.0 ; short-range electrostatic cutoff (in nm)
rvdw = 1.0 ; short-range van der Waals cutoff (in nm)
; Electrostatics
coulombtype = PME ; Particle Mesh Ewald for long-range electrostatics
pme_order = 4 ; cubic interpolation
fourierspacing = 0.16 ; grid spacing for FFT
; Temperature coupling is on
tcoupl = V-rescale ; modified Berendsen thermostat
tc-grps = Protein Non-Protein ; two coupling groups - more accurate
tau_t = 0.1 0.1 ; time constant, in ps
ref_t = 300 300 ; reference temperature, one for each group, in K
; Pressure coupling is on
pcoupl = Parrinello-Rahman ; Pressure coupling on in NPT
pcoupltype = isotropic ; uniform scaling of box vectors
tau_p = 2.0 ; time constant, in ps
ref_p = 1.0 ; reference pressure, in bar
compressibility = 4.5e-5 ; isothermal compressibility of water, bar^-1
; Periodic boundary conditions
pbc = xyz ; 3-D PBC
; Dispersion correction
DispCorr = EnerPres ; account for cut-off vdW scheme
; Velocity generation
gen_vel = no ; Velocity generation is off
2. 正式模拟步骤
目标:构建一个物理合理的模拟体系,为后续的动力学模拟奠定基础。
1. 加载分子结构(PDB 文件):
• PDB 文件(Protein Data Bank)包含分子(如蛋白质、核酸、小分子)的三维坐标和结构信息。
• GROMACS 使用 pdb2gmx 工具将这些坐标信息转化为分子模型,同时分配力场参数。
• 力场:描述分子间相互作用的数学模型,包括键、角、二面角势能及范德华力、电荷相互作用。
• 常见力场:GROMOS 、 AMBER 、 CHARMM 。
(常用的为 AMBER99SB protein,OPLS-AA/L all-atom force field,charm36 力场(需要自行下载配置),其他的力场过老,不适合发文章)
2. 生成拓扑文件:
• 拓扑文件(topol.top)定义了模拟体系中每个分子的力场参数,如原子类型、键的类型及其参数等。
• 它与坐标文件结合,形成分子动力学模拟的基础。
3. 运行 pdb2gmx,选择力场:15 回车
#删除水分子(PDB 文件中的 “HOH” 残基)
grep -v HOH 1aki.pdb > 1AKI_clean.pdb
gmx_mpi pdb2gmx -f 1AKI_clean.pdb -o 1AKI_processed.gro -water spce
#选择 15,按回车,OPLS-AA/L all-atom force field (2001 aminoacid dihedrals)
(base) root@liangzhong-4ay9ej85pxvd-main:/input0# pdb2gmx -f 1AKI_clean.pdb -o 1AKI_processed.gro -water spce
:-) GROMACS - gmx pdb2gmx, 2023 (-:
Executable: /data/app/gromacs/bin/gmx_mpi
Data prefix: /data/app/gromacs
Working dir: /input0
Command line:
gmx_mpi pdb2gmx -f 1AKI_clean.pdb -o 1AKI_processed.gro -water spce
Select the Force Field:
From '/data/app/gromacs/share/gromacs/top':
1: AMBER03 protein, nucleic AMBER94 (Duan et al., J. Comp. Chem. 24, 1999-2012, 2003)
2: AMBER94 force field (Cornell et al., JACS 117, 5179-5197, 1995)
3: AMBER96 protein, nucleic AMBER94 (Kollman et al., Acc. Chem. Res. 29, 461-469, 1996)
4: AMBER99 protein, nucleic AMBER94 (Wang et al., J. Comp. Chem. 21, 1049-1074, 2000)
5: AMBER99SB protein, nucleic AMBER94 (Hornak et al., Proteins 65, 712-725, 2006)
6: AMBER99SB-ILDN protein, nucleic AMBER94 (Lindorff-Larsen et al., Proteins 78, 1950-58, 2010)
7: AMBERGS force field (Garcia & Sanbonmatsu, PNAS 99, 2782-2787, 2002)
8: CHARMM27 all-atom force field (CHARM22 plus CMAP for proteins)
9: GROMOS96 43a1 force field
10: GROMOS96 43a2 force field (improved alkane dihedrals)
11: GROMOS96 45a3 force field (Schuler JCC 2001 22 1205)
12: GROMOS96 53a5 force field (JCC 2004 vol 25 pag 1656)
13: GROMOS96 53a6 force field (JCC 2004 vol 25 pag 1656)
14: GROMOS96 54a7 force field (Eur. Biophys. J. (2011), 40,, 843-856, DOI: 10.1007/s00249-011-0700-9)
15: OPLS-AA/L all-atom force field (2001 aminoacid dihedrals)
15
Using the Oplsaa force field in directory oplsaa.ff
going to rename oplsaa.ff/aminoacids.r2b
Opening force field file /data/app/gromacs/share/gromacs/top/oplsaa.ff/aminoacids.r2b
Reading 1AKI_clean.pdb...
WARNING: all CONECT records are ignored
Read 'LYSOZYME', 1001 atoms
Analyzing pdb file
Splitting chemical chains based on TER records or chain id changing.
There are 1 chains and 0 blocks of water and 129 residues with 1001 atoms
chain #res #atoms
1 'A' 129 1001
All occupancies are one
All occupancies are one
Opening force field file /data/app/gromacs/share/gromacs/top/oplsaa.ff/atomtypes.atp
Reading residue database... (Oplsaa)
Opening force field file /data/app/gromacs/share/gromacs/top/oplsaa.ff/aminoacids.rtp
Opening force field file /data/app/gromacs/share/gromacs/top/oplsaa.ff/aminoacids.hdb
Opening force field file /data/app/gromacs/share/gromacs/top/oplsaa.ff/aminoacids.n.tdb
Opening force field file /data/app/gromacs/share/gromacs/top/oplsaa.ff/aminoacids.c.tdb
Processing chain 1 'A' (1001 atoms, 129 residues)
Analysing hydrogen-bonding network for automated assignment of histidine
protonation. 213 donors and 184 acceptors were found.
There are 255 hydrogen bonds
Will use HISE for residue 15
Identified residue LYS1 as a starting terminus.
Identified residue LEU129 as a ending terminus.
8 out of 8 lines of specbond.dat converted successfully
Special Atom Distance matrix:
CYS6 MET12 HIS15 CYS30 CYS64 CYS76 CYS80
SG48 SD87 NE2118 SG238 SG513 SG601 SG630
MET12 SD87 1.166
HIS15 NE2118 1.776 1.019
CYS30 SG238 1.406 1.054 2.069
CYS64 SG513 2.835 1.794 1.789 2.241
CYS76 SG601 2.704 1.551 1.468 2.116 0.765
CYS80 SG630 2.959 1.951 1.916 2.391 0.199 0.944
CYS94 SG724 2.550 1.407 1.382 1.975 0.665 0.202 0.855
MET105 SD799 1.827 0.911 1.683 0.888 1.849 1.461 2.036
CYS115 SG889 1.576 1.084 2.078 0.200 2.111 1.989 2.262
CYS127 SG981 0.197 1.072 1.721 1.313 2.799 2.622 2.934
CYS94 MET105 CYS115
SG724 SD799 SG889
MET105 SD799 1.381
CYS115 SG889 1.853 0.790
CYS127 SG981 2.475 1.686 1.483
Linking CYS-6 SG-48 and CYS-127 SG-981...
Linking CYS-30 SG-238 and CYS-115 SG-889...
Linking CYS-64 SG-513 and CYS-80 SG-630...
Linking CYS-76 SG-601 and CYS-94 SG-724...
Start terminus LYS-1: NH3+
End terminus LEU-129: COO-
Checking for duplicate atoms....
Generating any missing hydrogen atoms and/or adding termini.
Now there are 129 residues with 1960 atoms
Making bonds...
Number of bonds was 1984, now 1984
Generating angles, dihedrals and pairs...
Before cleaning: 5142 pairs
Before cleaning: 5187 dihedrals
Making cmap torsions...
There are 5187 dihedrals, 426 impropers, 3547 angles
5106 pairs, 1984 bonds and 0 virtual sites
Total mass 14313.193 a.m.u.
Total charge 8.000 e
Writing topology
Writing coordinate file...
--------- PLEASE NOTE ------------
You have successfully generated a topology from: 1AKI_clean.pdb.
The Oplsaa force field and the spce water model are used.
--------- ETON ESAELP ------------
GROMACS reminds you: "Any one who considers arithmetical methods of producing random digits is, of course, in a state of sin." (John von Neumann)
可以看到多了几个文件,而且「Total charge 8.000 e」稍后我们需要中和电荷
使用 vim 命令查看 topol.top 可以看到多了力场标签
(base) root@liangzhong-4ay9ej85pxvd-main:/input0# ls
1AKI_clean.pdb 1aki.pdb md.mdp npt.mdp posre.itp
1AKI_processed.gro ions.mdp minim.mdp nvt.mdp topol.top
4. 创建菱形十二面体盒子,将蛋白质罩进去,以便我们添加溶剂分子
• 盒形选择:常见的模拟盒形状有立方体、正交盒、菱形十二面体等,通常选择能有效填充分子和减少计算量的形状。
gmx_mpi editconf -f 1AKI_processed.gro -o 1AKI_newbox.gro -c -d 1.0 -bt cubic
#将蛋白质在框中居中(-c),并将其放置在框边缘至少 1.0 nm 的位置(-d 1.0)
Command line:
gmx_mpi editconf -f 1AKI_processed.gro -o 1AKI_newbox.gro -c -d 1.0 -bt cubic
Note that major changes are planned in future for editconf, to improve usability and utility.
Read 1960 atoms
Volume: 123.376 nm^3, corresponds to roughly 55500 electrons
No velocities found
system size : 3.817 4.234 3.454 (nm)
diameter : 5.010 (nm)
center : 2.781 2.488 0.017 (nm)
box vectors : 5.906 6.845 3.052 (nm)
box angles : 90.00 90.00 90.00 (degrees)
box volume : 123.38 (nm^3)
shift : 0.724 1.017 3.488 (nm)
new center : 3.505 3.505 3.505 (nm)
new box vectors : 7.010 7.010 7.010 (nm)
new box angles : 90.00 90.00 90.00 (degrees)
new box volume : 344.48 (nm^3)
5. 定义了一个盒子,添加溶剂(水)
目的:溶剂化:将分子放入溶剂环境中(如水盒),以模拟其在实际环境中的行为。
gmx_mpi solvate -cp 1AKI_newbox.gro -cs spc216.gro -o 1AKI_solv.gro -p topol.top
#结果
Generating solvent configuration
Will generate new solvent configuration of 4x4x4 boxes
Solvent box contains 39252 atoms in 13084 residues
Removed 5451 solvent atoms due to solvent-solvent overlap
Removed 1869 solvent atoms due to solute-solvent overlap
Sorting configuration
Found 1 molecule type:
SOL ( 3 atoms): 10644 residues
Generated solvent containing 31932 atoms in 10644 residues
Writing generated configuration to 1AKI_solv.gro
Output configuration contains 33892 atoms in 10773 residues
Volume : 344.484 (nm^3)
Density : 997.935 (g/l)
Number of solvent molecules: 10644
Processing topology
Adding line for 10644 solvent molecules with resname (SOL) to topology file (topol.top)
#查看一下 top 文件
(base) root@liangzhong-4ay9ej85pxvd-main:/input0# ls
'#topol.top.1#' 1AKI_newbox.gro 1AKI_solv.gro ions.mdp minim.mdp nvt.mdp topol.top
1AKI_clean.pdb 1AKI_processed.gro 1aki.pdb md.mdp npt.mdp posre.itp
(base) root@liangzhong-4ay9ej85pxvd-main:/input0# vim topol.top
#查看 top 文件的末尾
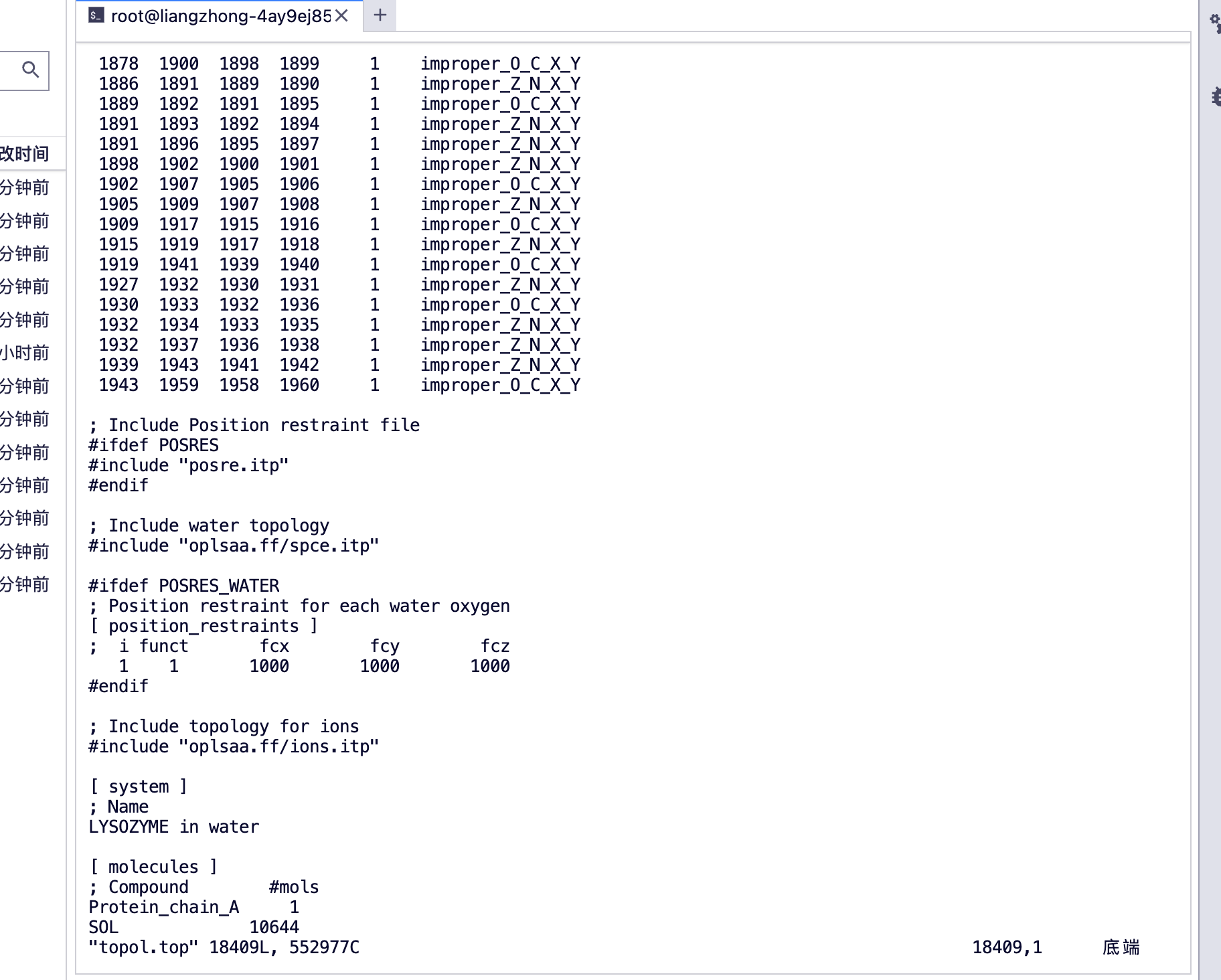
可以看到 [ molecules ]
protein_chain_A, 蛋白 A 链
SOL 水分子
6. 组装.tpr 文件
gmx_mpi grompp -f ions.mdp -c 1AKI_solv.gro -p topol.top -o ions.tpr
7. 中和电荷,用离子替代溶剂分子
电中性化:添加离子(如 Na⁺, Cl⁻)中和体系总电荷,避免因静电效应引起的模拟失真。
gmx_mpi genion -s ions.tpr -o 1AKI_solv_ions.gro -p topol.top -pname NA -nname CL -neutral
Command line:
gmx_mpi genion -s ions.tpr -o 1AKI_solv_ions.gro -p topol.top -pname NA -nname CL -neutral
Reading file ions.tpr, VERSION 2023 (single precision)
Reading file ions.tpr, VERSION 2023 (single precision)
Will try to add 0 NA ions and 8 CL ions.
Select a continuous group of solvent molecules
Group 0 ( System) has 33892 elements
Group 1 ( Protein) has 1960 elements
Group 2 ( Protein-H) has 1001 elements
Group 3 ( C-alpha) has 129 elements
Group 4 ( Backbone) has 387 elements
Group 5 ( MainChain) has 517 elements
Group 6 ( MainChain+Cb) has 634 elements
Group 7 ( MainChain+H) has 646 elements
Group 8 ( SideChain) has 1314 elements
Group 9 ( SideChain-H) has 484 elements
Group 10 ( Prot-Masses) has 1960 elements
Group 11 ( non-Protein) has 31932 elements
Group 12 ( Water) has 31932 elements
Group 13 ( SOL) has 31932 elements
Group 14 ( non-Water) has 1960 elements
Select a group: 13
Selected 13: 'SOL'
Number of (3-atomic) solvent molecules: 10644
Processing topology
Replacing 8 solute molecules in topology file (topol.top) by 0 NA and 8 CL ions.
Back Off! I just backed up topol.top to ./#topol.top.2#
Using random seed -1212354563.
Replacing solvent molecule 3671 (atom 12973) with CL
Replacing solvent molecule 2264 (atom 8752) with CL
Replacing solvent molecule 2559 (atom 9637) with CL
Replacing solvent molecule 8081 (atom 26203) with CL
Replacing solvent molecule 8468 (atom 27364) with CL
Replacing solvent molecule 7439 (atom 24277) with CL
Replacing solvent molecule 9983 (atom 31909) with CL
Replacing solvent molecule 650 (atom 3910) with CL
GROMACS reminds you: "Water is just water" (Berk Hess)
(base) root@liangzhong-4ay9ej85pxvd-main:/input0#
可以看到添加了很多氯离子
8. 能量最小化
8.1 原因:我们在进行分子动力学模拟的时候,由于我们获得的 pdb 文件是通过 x 射线,电镜等方法得到的,其中含有很多能量过高的键角、扭曲等,又或是氢键的形成或断裂,分子间的特殊排列,也可能由于原子之间距离太近造成的高能,使得模拟中的分子系统难以从高能状态向低能状态转变。此时需要使用这种方法使得体系达到能量最小化
8.2 原理:能量最小化的基本原理:能量最小化基于势能函数,通过迭代计算和调整原子坐标来减少系统的总势能。常用的方法包括:
①共轭梯度法(Conjugate Gradient Method):一种梯度下降法,通过利用前一步的信息来加速收敛。
②最速下降法(Steepest Descent Method):每次迭代沿着势能梯度的反方向移动,以最陡峭的下降路径减少能量。
③牛顿-拉弗森法(Newton-Raphson Method):一种二阶导数法,利用势能函数的二阶导数来更准确地找到最小能量点。
8.3 目的: 在通过调整分子的原子坐标来减少系统的总势能,从而找到一个更稳定的分子构象。
①去除不合理的几何构象:通过能量最小化,能够去除由于初始结构中的不合理几何构象,如不合理发原子重叠和拉伸,消除其所引起的高能量状态,使系统更加稳定。
②准备初始结构:确保初始结构处于较低的势能状态,以便模拟过程中避免不必要的高能量冲击。
③改善计算效率:通过减少系统的总势能,可以提高后续模拟和计算的稳定性和效率
gmx_mpi grompp -f minim.mdp -c 1AKI_solv_ions.gro -p topol.top -o em.tpr
gmx_mpi mdrun -v -deffnm em

9. 看一下结果(在教程的最后会统一教大家可视化 xvg 文件,可以用 xmgrace,qtgrace,python,excel 做图,本质上是个折线图)
gmx_mpi energy -f em.edr -o potential.xvg
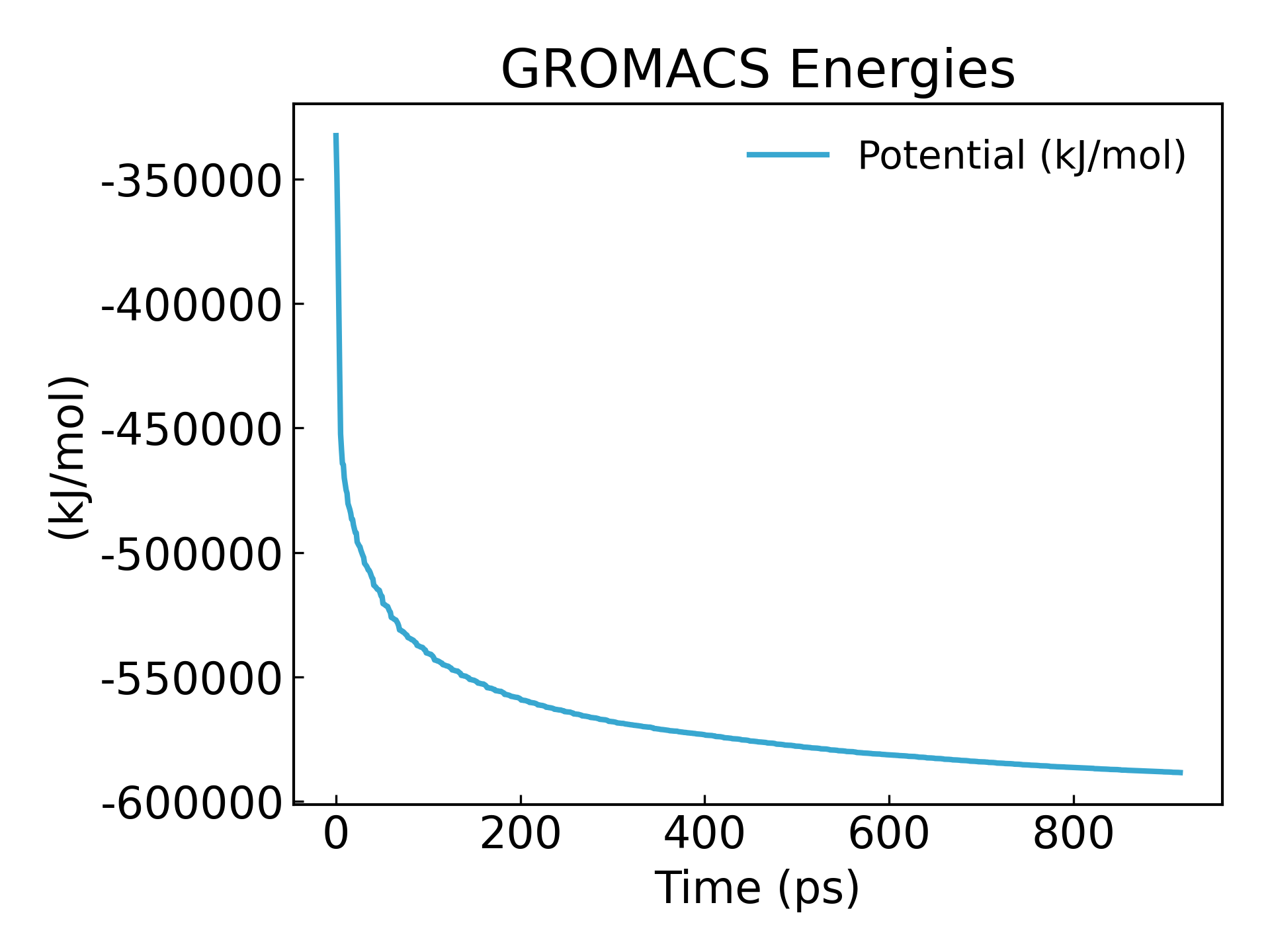
可以看到能量达到最小化了 -600000kj/mol
10. 体系平衡
目标:将体系调整到目标温度和压力条件下,使其接近真实物理状态。
(1)NVT 模拟(恒体积恒温):
• 恒体积恒温系综用于将体系温度稳定到目标值。
• 温度控制器:常用的是 Berendsen 温度耦合器或 V-rescale(修正的弱耦合方法),通过调整原子速度控制温度。
(2) NPT 模拟(恒压恒温):
• 恒压恒温系综进一步将体系密度调整到目标值(通常为液态水的密度 ~1 g/cm³)。
• 压力控制器:常用的是 Berendsen 压力耦合器或 Parrinello-Rahman 压力控制方法。
10.1. 进行 100ps 的 NVT 平衡,在(恒定粒子数、体积和温度)下进行的,也被称为「等温等容」
GPU 加速下非常快,只需要 10+ 秒。
gmx_mpi grompp -f nvt.mdp -c em.gro -r em.gro -p topol.top -o nvt.tpr
gmx_mpi mdrun -deffnm nvt -nb gpu -pme cpu
#将 PME 任务移至 CPU

gmx_mpi energy -f nvt.edr -o temperature.xvg
#生成温度随时间变化的图像,查看温度是否平衡
16
0
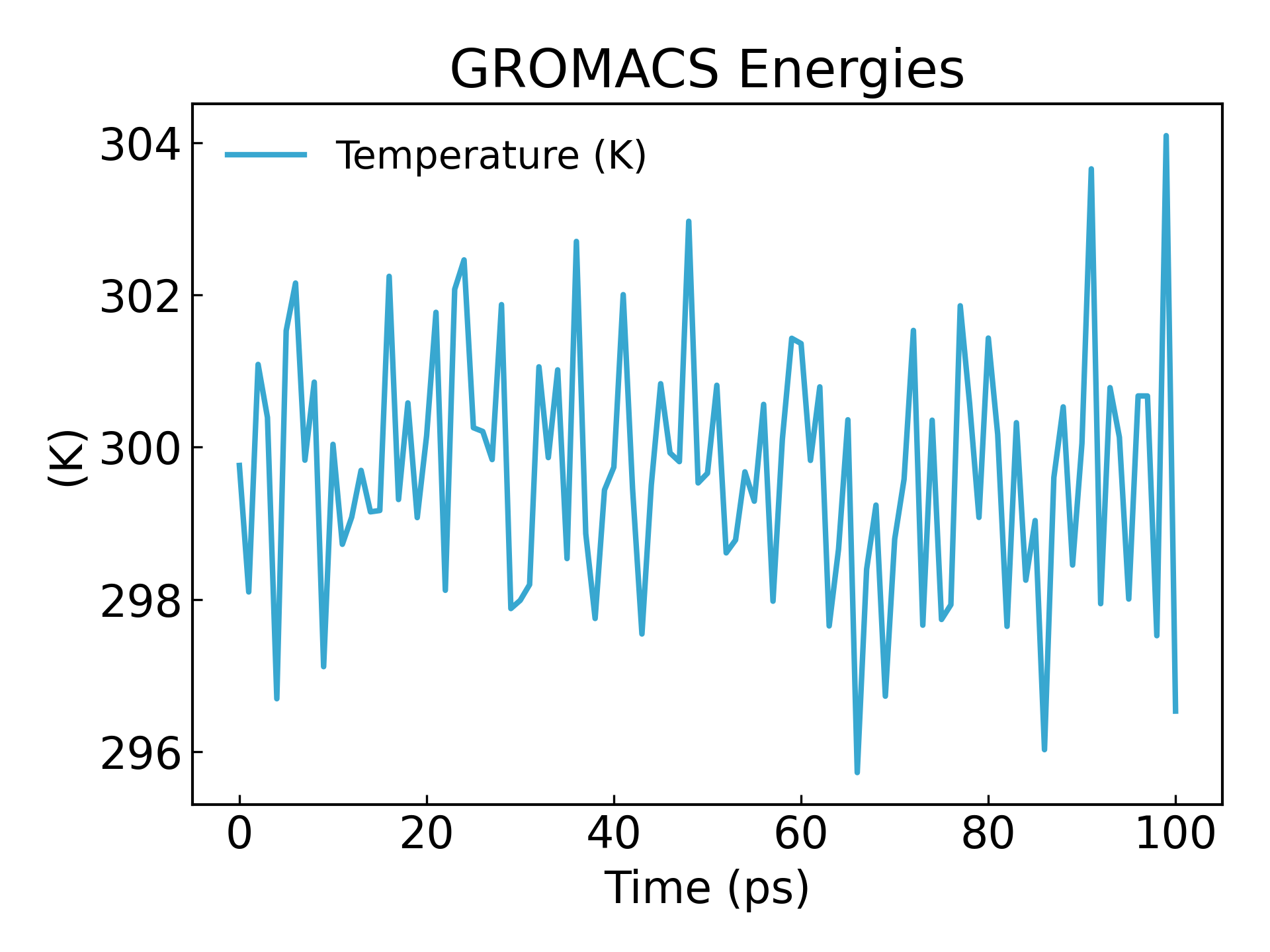
可以看到温度也在 100ps 内达到基本稳态
10.2.npt 等温等压” 平衡,稳定系统的压力,进行 100 ps NPT 平衡。
gmx_mpi grompp -f npt.mdp -c nvt.gro -r nvt.gro -t nvt.cpt -p topol.top -o npt.tpr
gmx_mpi mdrun -deffnm npt -nb gpu -pme cpu
#压力是否平衡
gmx_mpi energy -f npt.edr -o pressure.xvg
18
0
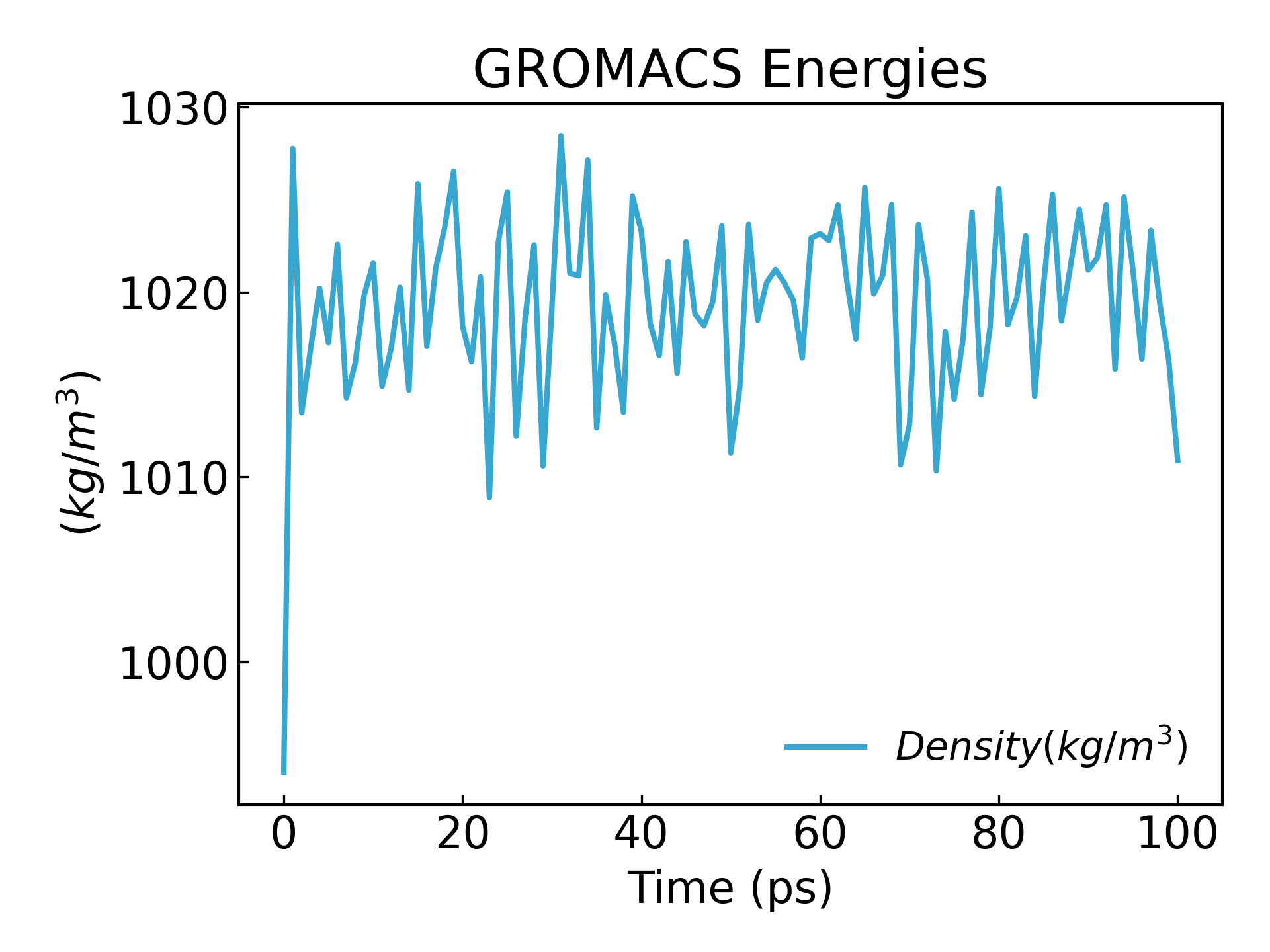
gmx_mpi energy -f npt.edr -o density.xvg
24
0
可以看到密度达到稳态:
11. 完成 2 个平衡阶段后,系统现在在所需的温度和压力下已良好平衡。我们现在可以释放位置约束并运行 MD 进行。
这里可以按照自己需求修改时间即可,本教程模拟 100ns 。
时间步长 dt = 2 fs(常见设置):
50000000 步,时间步长为 2 fs,相当于 100 纳秒 (ns)
50000000 x2 fs = 10^8 fs = 10^5 ps =100ns
vim md.mdp
nsteps = 50000000 ; 2 * 50000000 = 100000 ps (100 ns)

gmx_mpi grompp -f md.mdp -c npt.gro -t npt.cpt -p topol.top -o md_0_1.tpr
##最后一步提交,pme 分配到 CPU 上:
gmx_mpi mdrun -deffnm md_0_1 -v -nb gpu -pme cpu
-v 可以显示运行剩余的时间。
3. 结果分析
1.trjconv,它被用作后处理工具,用于去除坐标、校正周期性或手动修改轨迹(时间单位、帧频率等),这是因为在所有带有周期性边界条件的模拟中,分子都有可能出现断裂 (broken) 或者在盒子边缘跳来跳去 (jump) 的情况,可以把分子重新放在盒子中间,rewrap 分子,并且恢复盒子的菱形十二面体形状 (rhombic dodecahedral shape) 。
-pbc mol: 去除轨迹中的周期性边界条件 (PBC),并基于分子进行修正(即确保每个分子在轨迹中是完整的)。
• -center: 将选定的分子/分组居中到模拟框的中心。
gmx_mpi trjconv -s md_0_1.tpr -f md_0_1.xtc -o md_0_1_noPBC.xtc -pbc mol -center
Select group for centering
Group 0 ( System) has 33876 elements
Group 1 ( Protein) has 1960 elements
Group 2 ( Protein-H) has 1001 elements
Group 3 ( C-alpha) has 129 elements
Group 4 ( Backbone) has 387 elements
Group 5 ( MainChain) has 517 elements
Group 6 ( MainChain+Cb) has 634 elements
Group 7 ( MainChain+H) has 646 elements
Group 8 ( SideChain) has 1314 elements
Group 9 ( SideChain-H) has 484 elements
Group 10 ( Prot-Masses) has 1960 elements
Group 11 ( non-Protein) has 31916 elements
Group 12 ( Water) has 31908 elements
Group 13 ( SOL) has 31908 elements
Group 14 ( non-Water) has 1968 elements
Group 15 ( Ion) has 8 elements
Group 16 ( Water_and_ions) has 31916 elements
Select a group: 1
Selected 1: 'Protein'
Select group for output
Group 0 ( System) has 33876 elements
Group 1 ( Protein) has 1960 elements
Group 2 ( Protein-H) has 1001 elements
Group 3 ( C-alpha) has 129 elements
Group 4 ( Backbone) has 387 elements
Group 5 ( MainChain) has 517 elements
Group 6 ( MainChain+Cb) has 634 elements
Group 7 ( MainChain+H) has 646 elements
Group 8 ( SideChain) has 1314 elements
Group 9 ( SideChain-H) has 484 elements
Group 10 ( Prot-Masses) has 1960 elements
Group 11 ( non-Protein) has 31916 elements
Group 12 ( Water) has 31908 elements
Group 13 ( SOL) has 31908 elements
Group 14 ( non-Water) has 1968 elements
Group 15 ( Ion) has 8 elements
Group 16 ( Water_and_ions) has 31916 elements
Select a group: 0
Selected 0: 'System'
选择 4(「骨架」)用于最小二乘拟合和 RMSD 计算。
2.RMSD
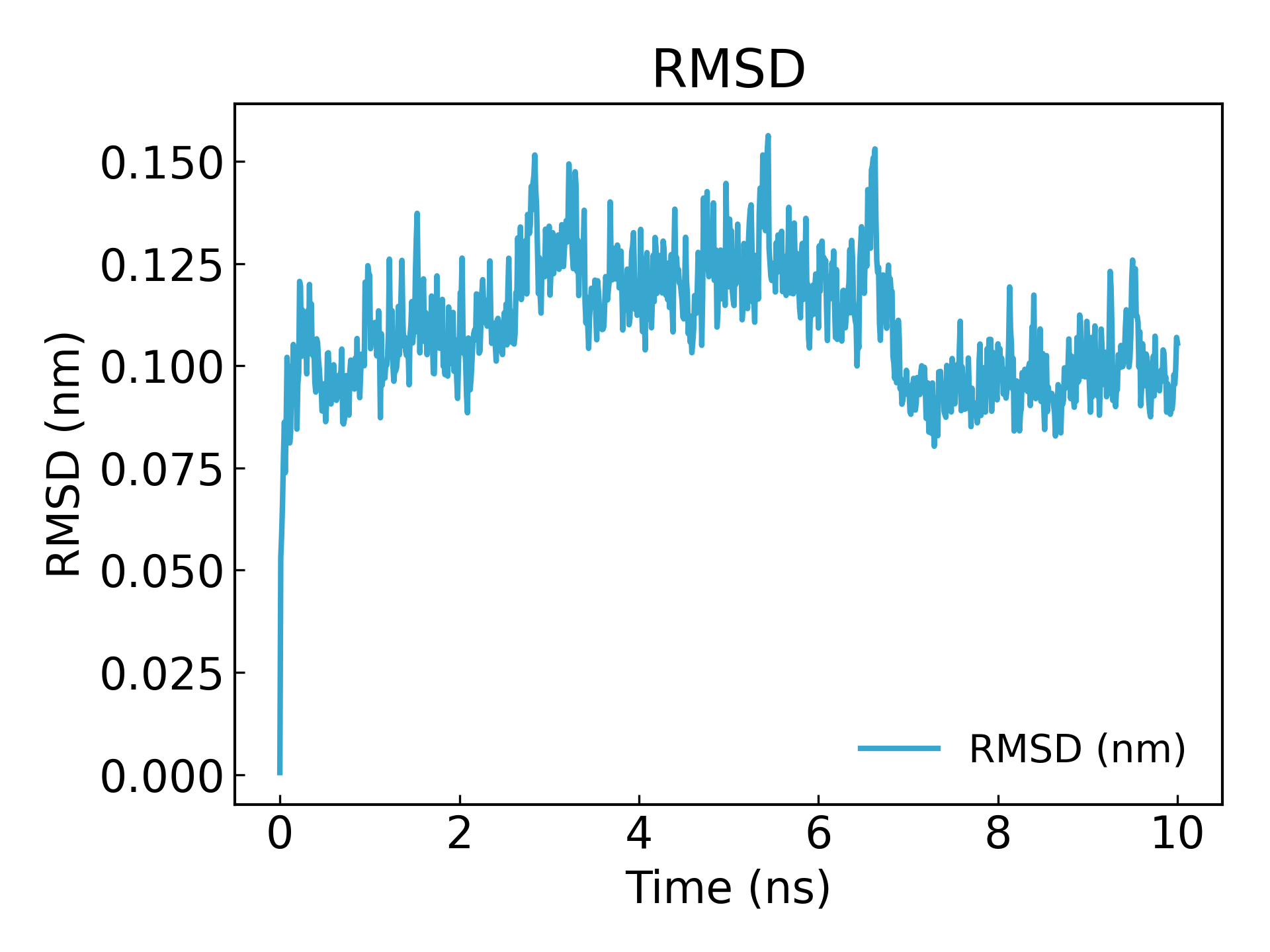
可用于查看模拟的收敛性和蛋白的稳定性. 用**g_rms**计算模拟过程中的结构与初始结构的 RMSD, 某时刻结构相对于 0ns 的偏差,常常用来评估蛋白结构的稳定性,在药物设计领域经常用到,如蛋白配体结构的稳定性,RMSD 曲线的波动幅度较小,且平稳说明配体和受体亲和力很大
gmx_mpi rms -s md_0_1.tpr -f md_0_1_noPBC.xtc -o rmsd.xvg
选择 4(「骨架」)用于最小二乘拟合和 RMSD 计算
可以看到在 10ns 后达到了平衡(注意:在发表或投稿文章时最好模拟时间达到 100ns,或者 50ns)
3. 回旋半径分析
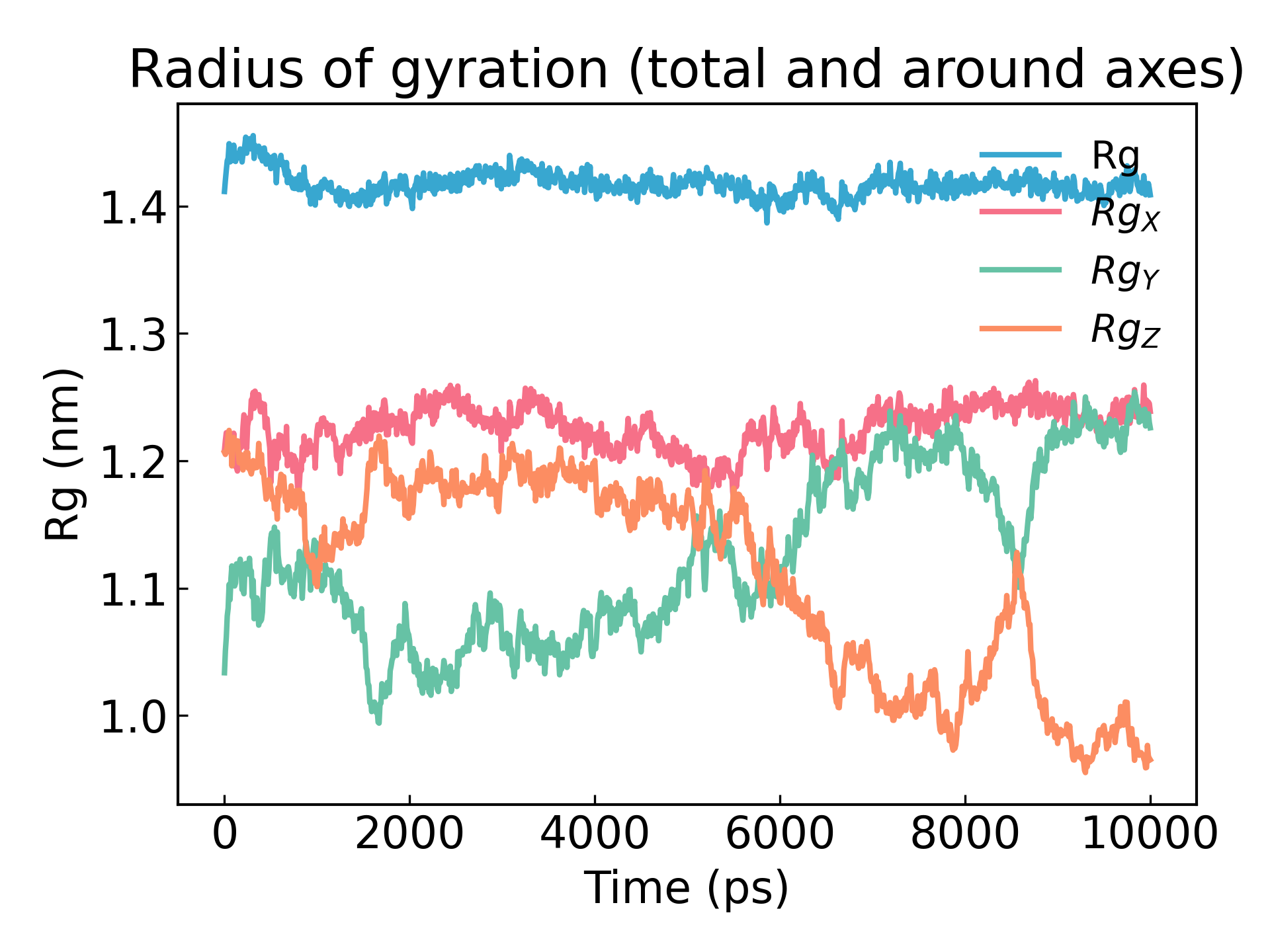
gmx_mpi gyrate -s md_0_1.tpr -f md_0_1_noPBC.xtc -o gyrate.xvg
蛋白质的回转半径是其紧密度的度量。如果一个蛋白质结构稳定,它将可能保持一个相对稳定的 R g 值。如果一个蛋白质展开,其 R g 值将随时间变化。让我们分析我们模拟中溶菌酶的回转半径:
4. 可视化作图
推荐软件:DuIvyTools:GROMACS 模拟分析与可视化工具
来自:https://github.com/CharlesHahn/DuIvyTools
pip install DuIvyTools
dit xvg_show -f rmsd.xvg -o rmsd_plot.png
dit xvg_show -f rmsd.xvg -o rmsd_plot.png
dit xvg_show -f potential.xvg -o potential_plot.png
dit xvg_show -f temperature.xvg -o temperature_plot.png
dit xvg_show -f density.xvg -o density_plot.png
dit xvg_show -f pressure.xvg -o pressure_plot.png
打开我们自己创建的数据集
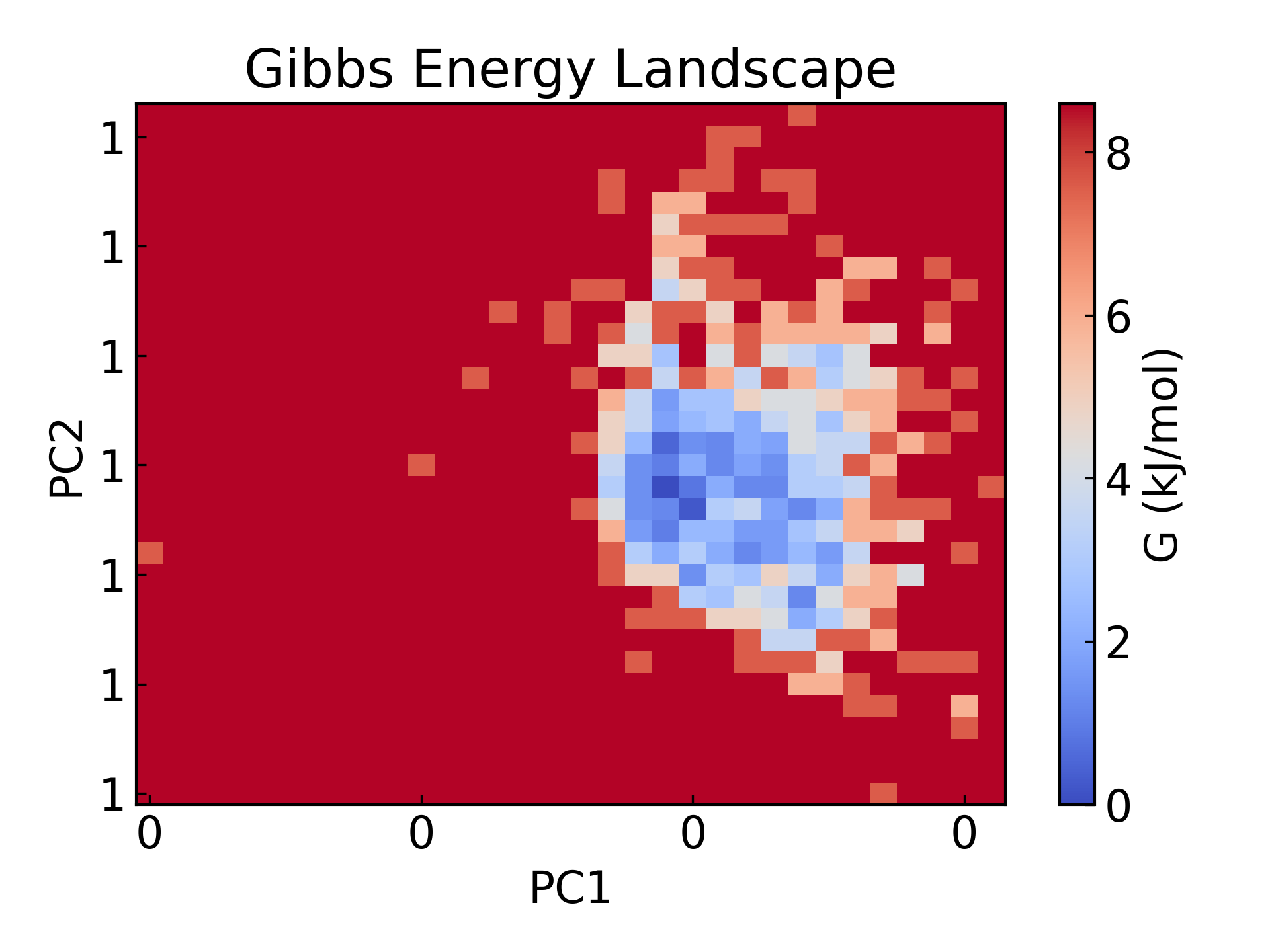
5.GROMACS 绘制能量景观图自由能形貌图绘制,将回旋半径和 rmsd 分别作为两个 PCA 成分。
gmx_mpi gyrate -s md_0_1.tpr -f md_0_1_noPBC.xtc -o rg.xvg
gmx_mpi rms -s md_0_1.tpr -f md_0_1_noPBC.xtc -o rmsd.xvg
vim rmsd.xvg
#输入以下命令删除以 # 或 @ 开头的行:
:g/^[@#]/d
vim rg.xvg
#输入以下命令删除以 # 或 @ 开头的行:
:g/^[@#]/d
#注意不要有空行,
paste rmsd.xvg rg.xvg > rmsd-rg.xvg
(base) # tail -f rmsd-rg.xvg
9910.0000000 0.0925907 9910 1.37624 1.20925 1.20765 0.931324
9920.0000000 0.0881348 9920 1.38369 1.22248 1.21078 0.932077
9930.0000000 0.0911074 9930 1.39224 1.23799 1.21709 0.928842
9940.0000000 0.0893596 9940 1.38188 1.21942 1.21672 0.922916
9950.0000000 0.0915931 9950 1.37509 1.21939 1.20194 0.922051
9960.0000000 0.0978161 9960 1.38113 1.2262 1.21084 0.919414
9970.0000000 0.0954911 9970 1.37934 1.21241 1.20711 0.937075
9980.0000000 0.0993617 9980 1.38301 1.22353 1.21083 0.92862
9990.0000000 0.1069279 9990 1.37943 1.22317 1.20579 0.924978
10000.0000000 0.1055321 10000 1.37524 1.21648 1.20194 0.92632
^Z
[11]+ Stopped tail -f rmsd-rg.xvg
#查看已经将两个文件整合在一起了,我们只要保留
#从 rmsd-rg.xvg 文件内容可以看到,每一行的数据格式如下:
时间_RMSD(ns) RMSD 时间_Rg(ps) Rg 其他列
(base) tail -f rmsd.xvg
9910.0000000 0.0925907
9920.0000000 0.0881348
9930.0000000 0.0911074
9940.0000000 0.0893596
9950.0000000 0.0915931
9960.0000000 0.0978161
9970.0000000 0.0954911
9980.0000000 0.0993617
9990.0000000 0.1069279
10000.0000000 0.1055321
^Z
[12]+ Stopped tail -f rmsd.xvg
#整理数据为
时间 (ns) RMSD Rg
(base) python
Python 3.8.15 (default, Nov 24 2022, 15:19:38)
[GCC 11.2.0] :: Anaconda, Inc. on linux
Type "help", "copyright", "credits" or "license" for more information.
# 加载数据
>>> import pandas as pd
>>> data = pd.read_csv("rmsd-rg.xvg", delim_whitespace=True, header=None, comment="#")
# 保留需要的列:第 1 列 (RMSD 时间) 、第 2 列 (RMSD) 、第 4 列 (Rg)
>>> cleaned_data = data[[0, 1, 3]]
# 将列名修改为更直观的名称
>>> cleaned_data.columns = ["Time (ps)", "RMSD", "Rg"]
>>> cleaned_data.to_csv("rmsd-rg-cleaned.xvg", sep="\t", index=False, header=False)
>>> exit()
#整理成功
tail -f rmsd-rg-cleaned.xvg
9910.0 0.0925907 1.37624
9920.0 0.0881348 1.38369
9930.0 0.0911074 1.39224
9940.0 0.0893596 1.38188
9950.0 0.0915931 1.37509
9960.0 0.0978161 1.38113
9970.0 0.0954911 1.37934
9980.0 0.0993617 1.38301
9990.0 0.1069279 1.37943
10000.0 0.1055321 1.37524
gmx_mpi sham -tsham 300 -nlevels 100 -f rmsd-rg-cleaned.xvg -ls gibbs.xpm -g gibbs.log -lsh enthalpy.xpm -lss entropy.xpm
dit xpm_show -f gibbs.xpm -o gibbs_2d.png
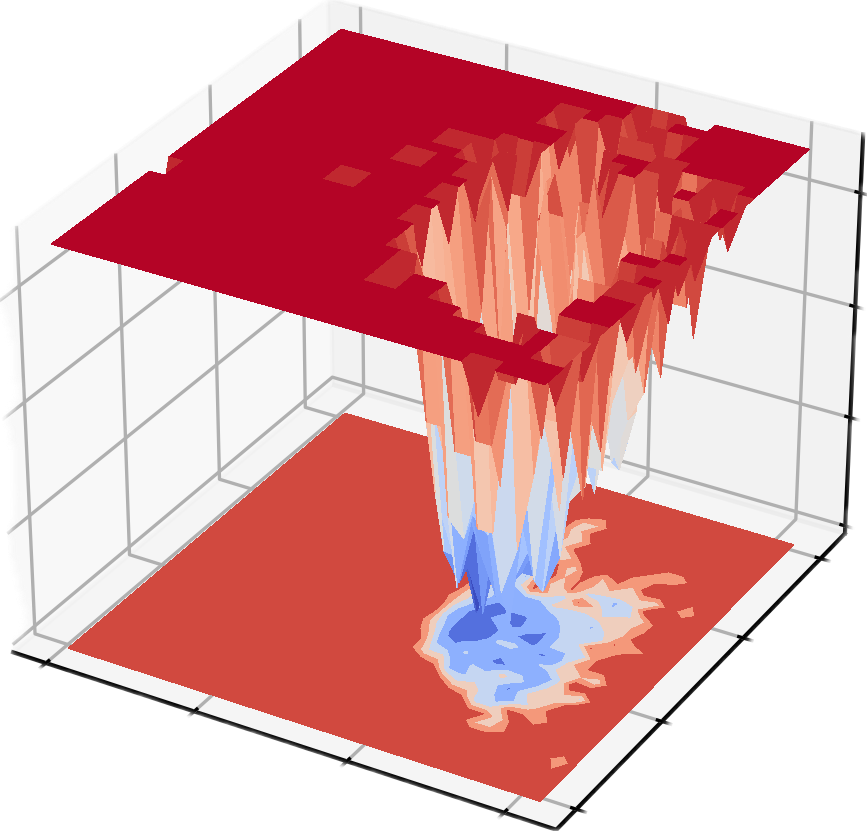
dit xpm_show -f gibbs.xpm -m 3d -o gibbs_3d.png
#tsham : 设定温度
#-nlevels: 设定 FEL 的层次数量
自由能形貌图 (Free Energy Landscape, FES) 是分子模拟中非常重要的工具,主要用于描述分子体系在特定坐标上的自由能分布。它的核心作用是揭示体系的热力学稳定性和动力学过程,通常在以下科研领域有重要意义:
- 稳态与过渡态研究:揭示体系的稳定构象(自由能最低点)和构象转换路径(自由能能垒),用于分析蛋白质折叠、化学反应和分子识别过程。
- 动力学路径与热力学稳定性:量化不同状态间的自由能差,帮助理解分子间的相互作用和构象转变的能量驱动。
- 药物设计与催化研究:预测配体-受体结合路径、能量障碍以及反应速率,为分子机制解析和功能优化提供理论指导。
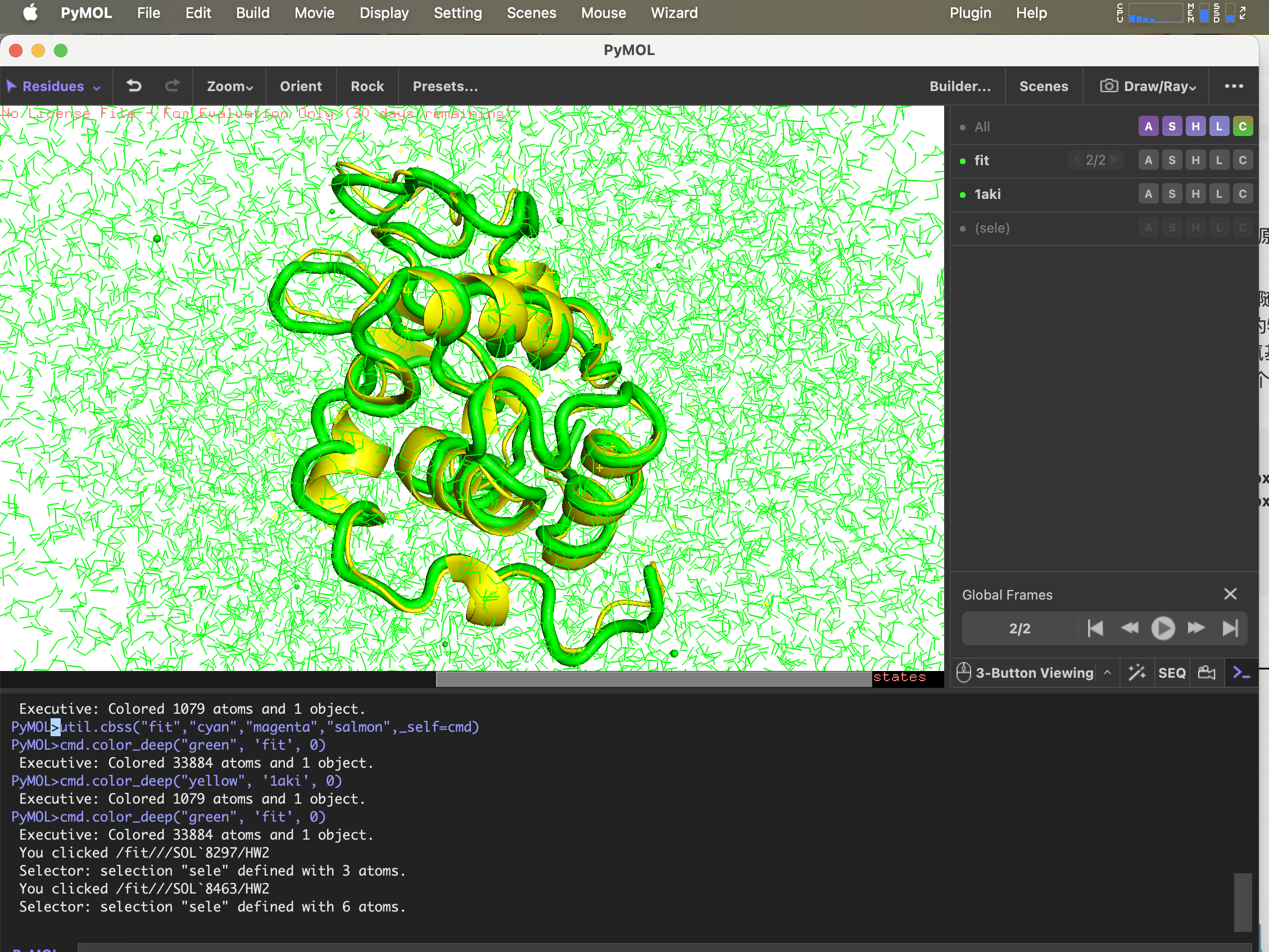
6.g_confrms 比较构象差异
要比较模拟完成后的结构与初始 PDB 文件中结构的差异,fit.pdb 包含两个分子结构的文件,两次都选择 4 (Backbone)
gmx_mpi confrms -f1 1AKI_clean.pdb -f2 md_0_1.gro -o fit.pdb
pdb 结构用 pymol 打开
7.g_rmsf 计算根均方涨落 RMSF,计算后 500 ps 范围内的平均结构,g_rmsf 得到的是随原子序号变化的曲线,选择 1 Protein
氨基酸残基的波动可以通过 RMSF 参数来推断,该参数解释了每个氨基酸残基/每个原子随时间的平均偏离参考位置的情况。更确切地说,它可以说是分析蛋白质结构中从其平均结构波动的特定部分。 RMSF 值高的氨基酸或氨基酸基团表明该配合物具有较大的柔韧性,而 RMSF 值较低的氨基酸则表明该络合物的柔韧性较差。频繁的波动稳定性更差 RMSF 值是一个动态参数,用于测量每个残基位置的平均主链柔性 [19],
gmx_mpi rmsf -s md_0_1.tpr -f md_0_1_noPBC.xtc -b 500 -o fws-rmsf.xvg -ox fws-avg.pdb
gmx_mpi rmsf -s md_0_1.tpr -f md_0_1_noPBC.xtc -b 500 -o fws-rmsf.xvg -ox fws-avg.pdb -res
dit xvg_show -f fws-rmsf.xvg -o rmsf_res.png
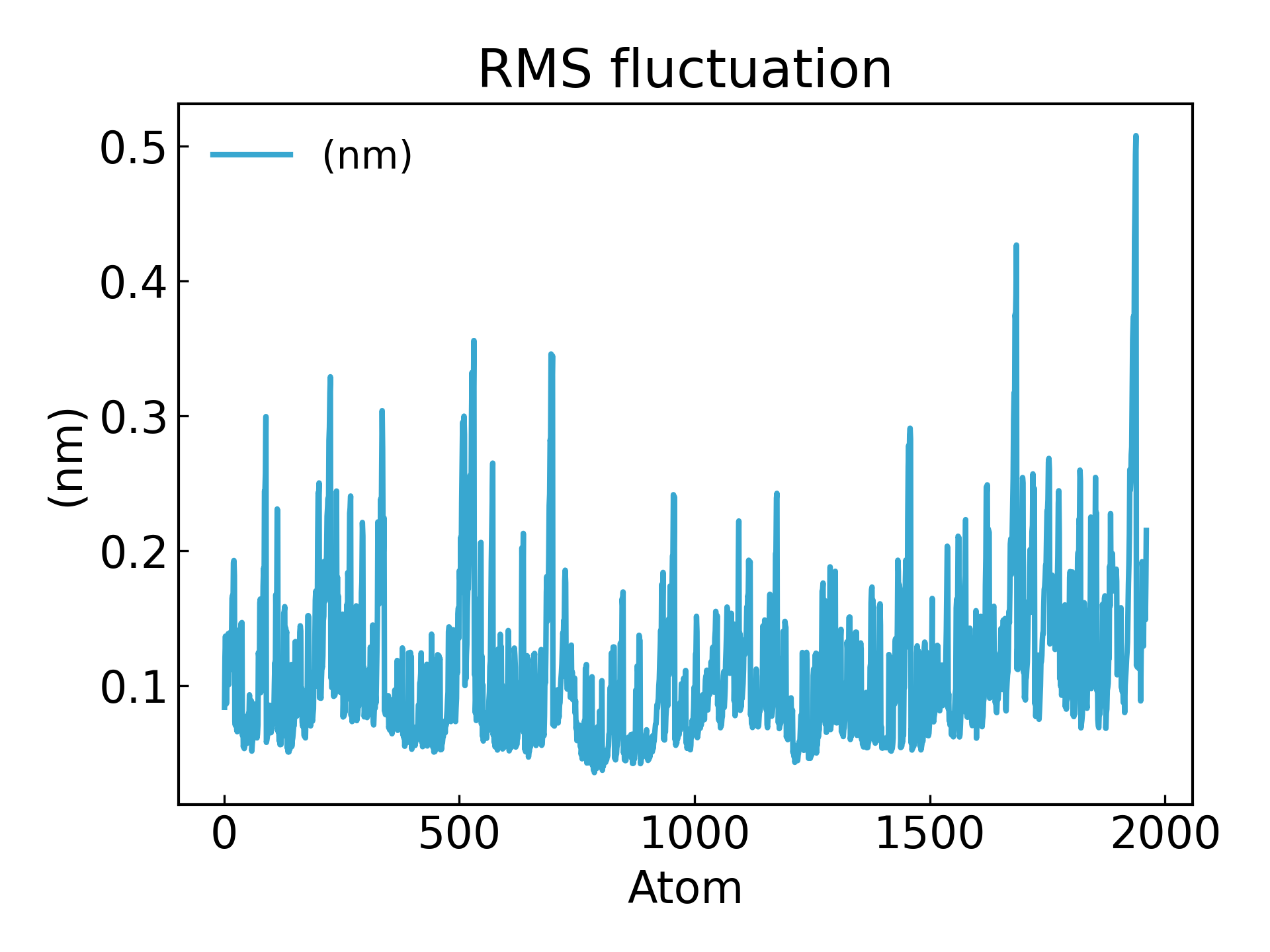
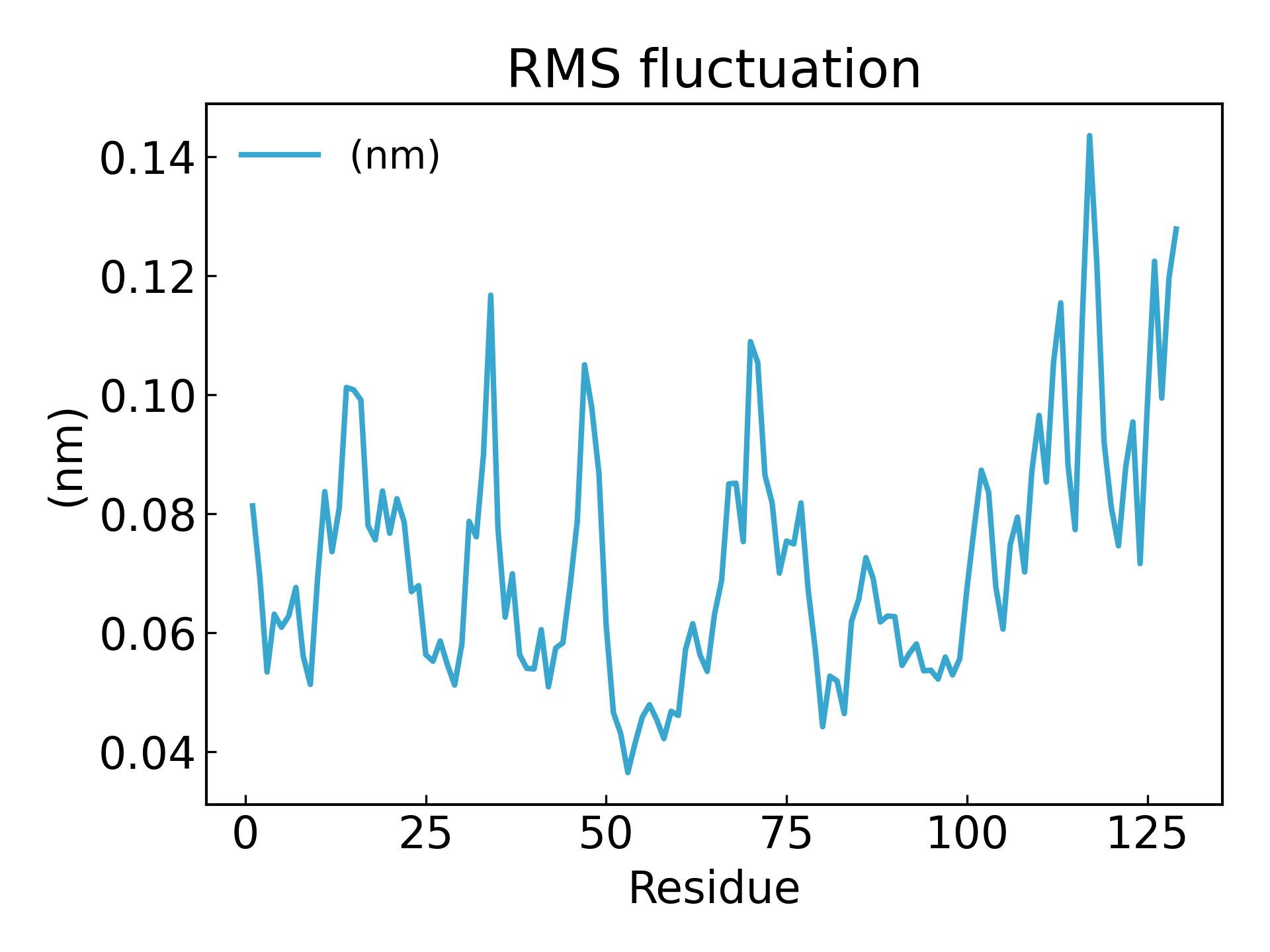
# 最后可以放到 pymol 里面看轨迹动画,点击播放即可
gmx_mpi trjconv -s md_0_1.tpr -f md_0_1_noPBC.xtc -o trajectory.pdb -skip 10